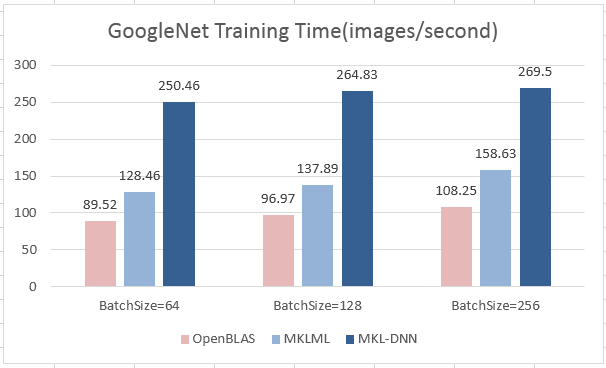
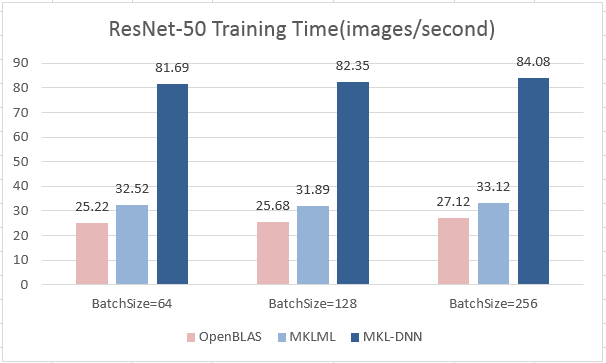
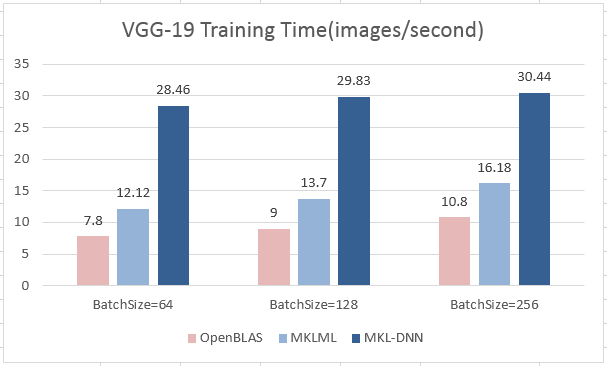
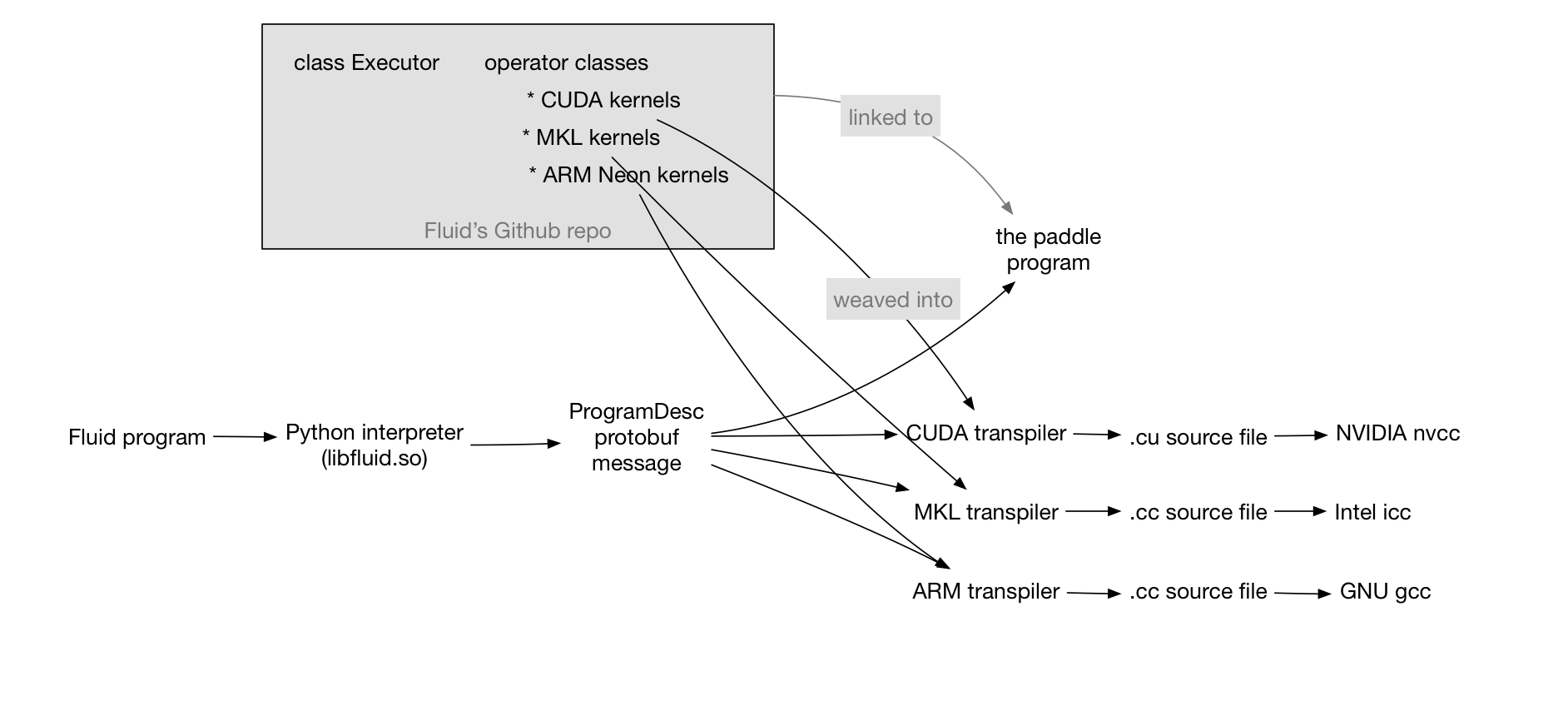
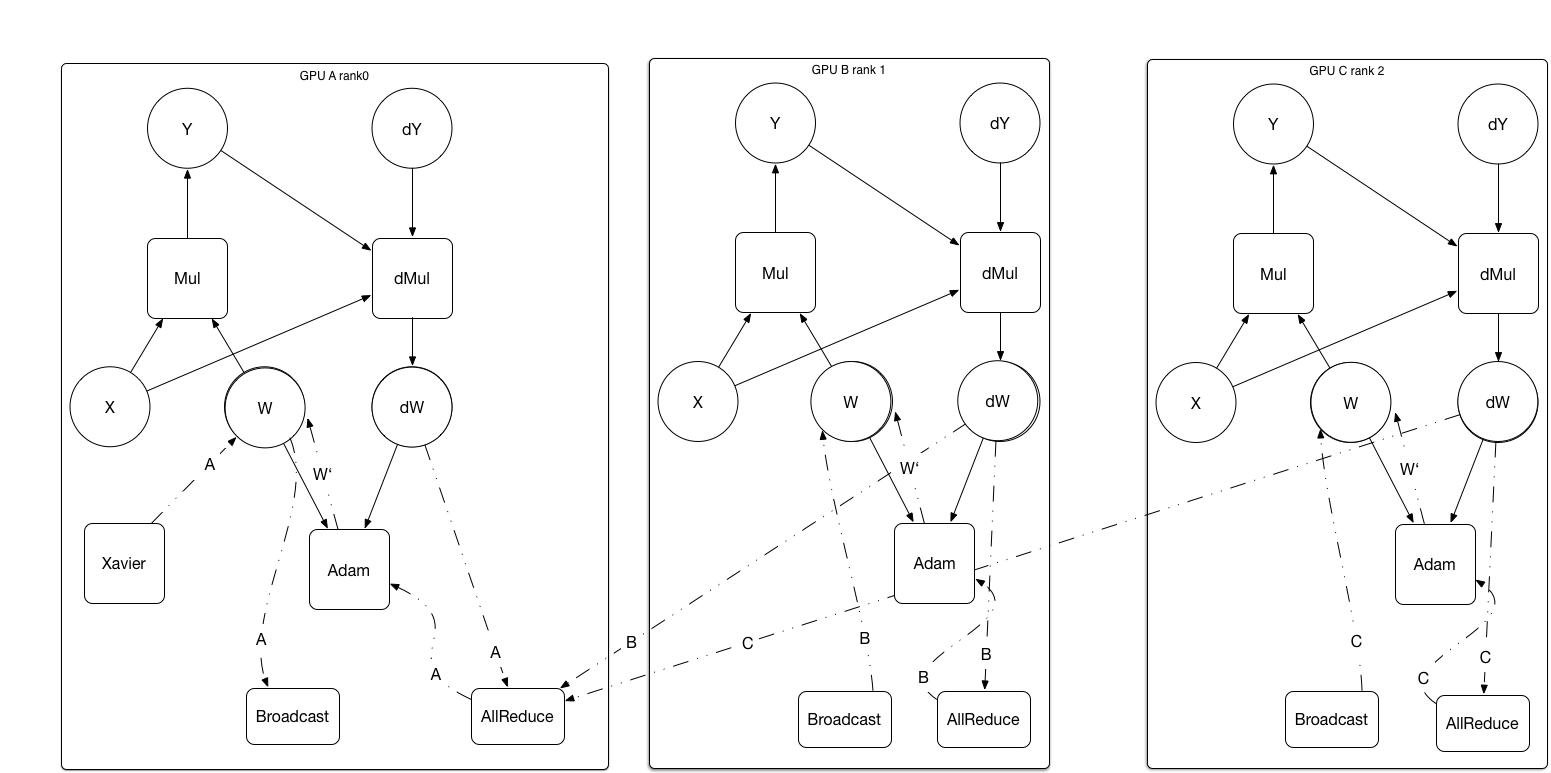
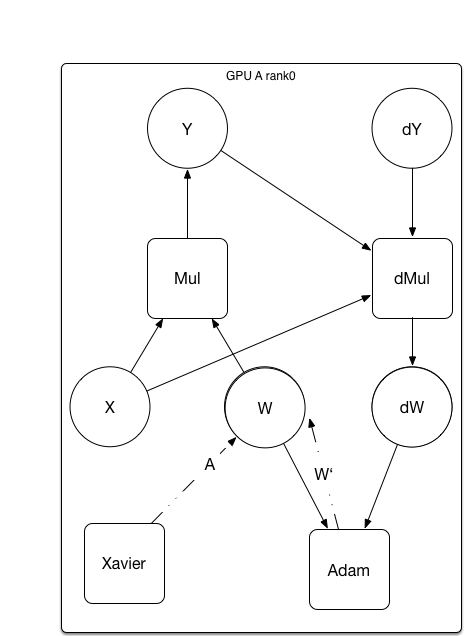
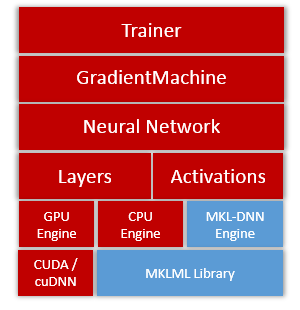
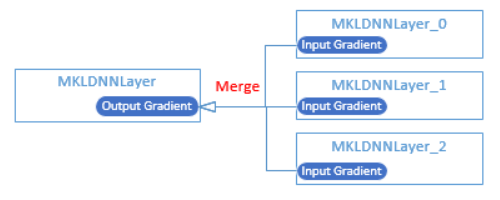
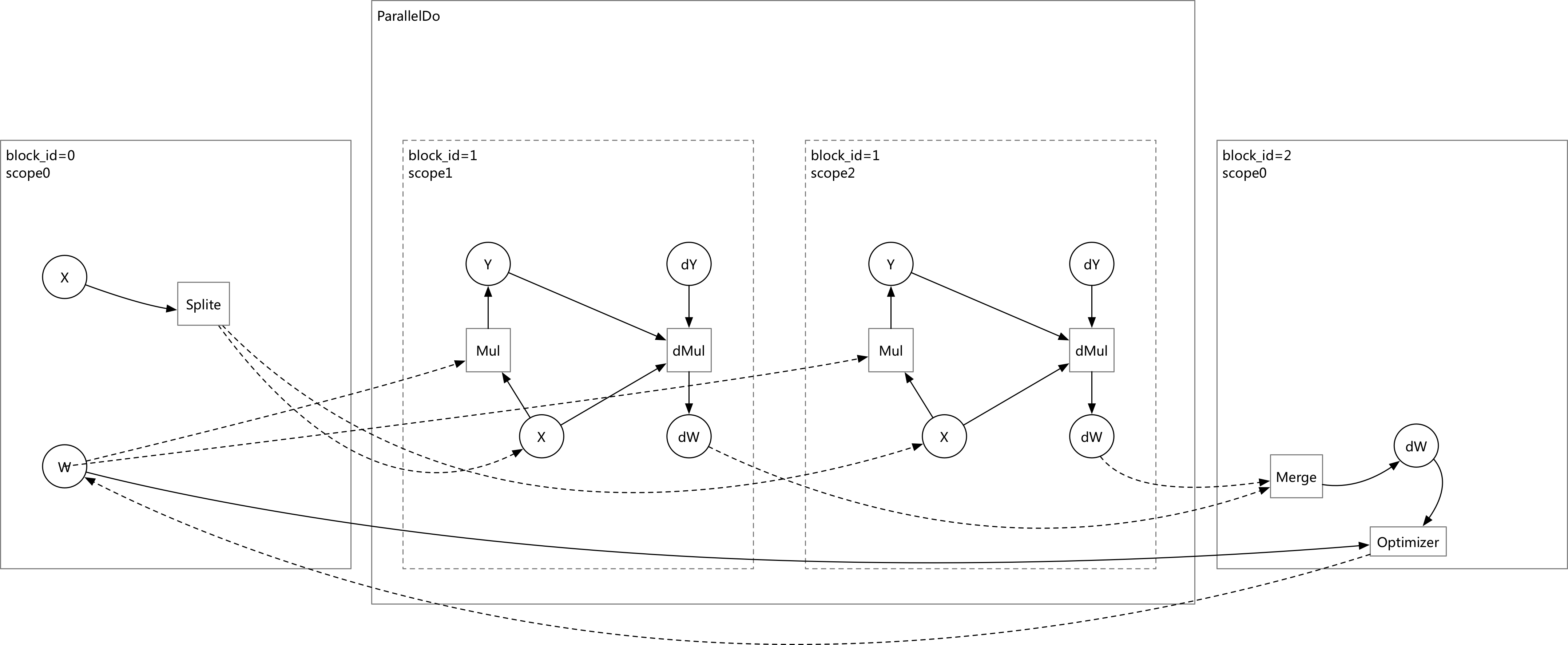
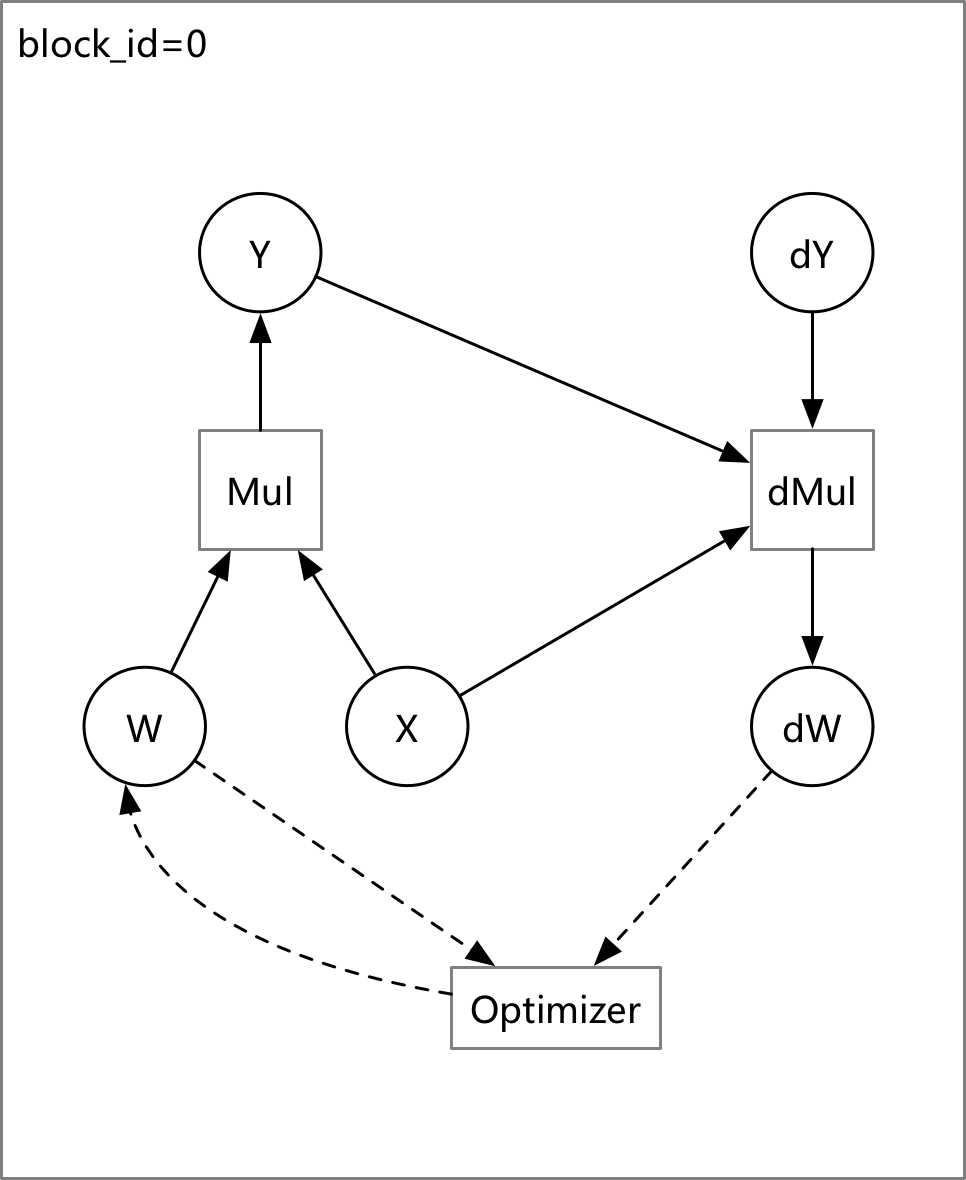
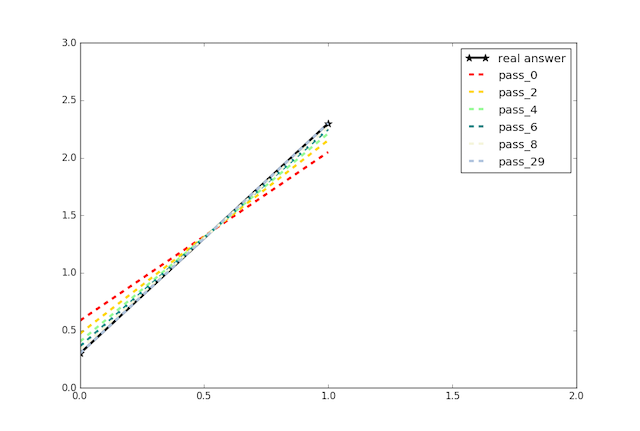
update design doc
Showing
17.8 KB
19.8 KB
benchmark/figs/vgg-cpu-train.png
0 → 100644
17.9 KB
cmake/external/cares.cmake
0 → 100644
cmake/external/grpc.cmake
0 → 100644
doc/api/v2/fluid.rst
0 → 100644
doc/api/v2/fluid/data_feeder.rst
0 → 100644
doc/api/v2/fluid/evaluator.rst
0 → 100644
doc/api/v2/fluid/executor.rst
0 → 100644
doc/api/v2/fluid/initializer.rst
0 → 100644
doc/api/v2/fluid/layers.rst
0 → 100644
doc/api/v2/fluid/nets.rst
0 → 100644
doc/api/v2/fluid/optimizer.rst
0 → 100644
doc/api/v2/fluid/param_attr.rst
0 → 100644
doc/api/v2/fluid/profiler.rst
0 → 100644
doc/api/v2/fluid/regularizer.rst
0 → 100644
doc/design/backward.md
0 → 100644
doc/design/fluid-compiler.graffle
0 → 100644
文件已添加
doc/design/fluid-compiler.png
0 → 100644
121.2 KB
doc/design/fluid.md
0 → 100644
文件已移动
文件已移动
文件已移动
文件已添加
108.4 KB
文件已添加
32.8 KB
doc/design/kernel_hint_design.md
0 → 100644
doc/design/mkl/image/engine.png
0 → 100644
13.3 KB
22.4 KB
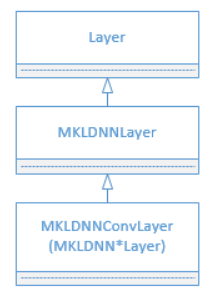
doc/design/mkl/image/layers.png
0 → 100644
11.4 KB
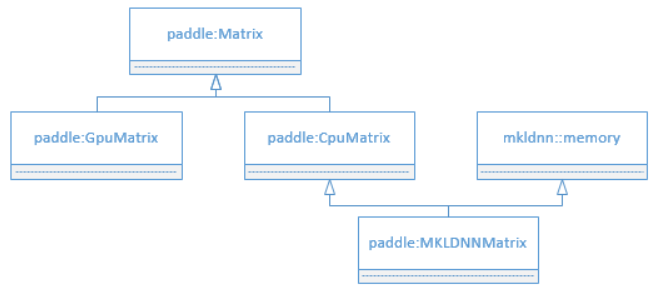
doc/design/mkl/image/matrix.png
0 → 100644
18.0 KB
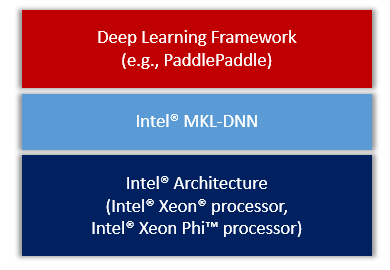
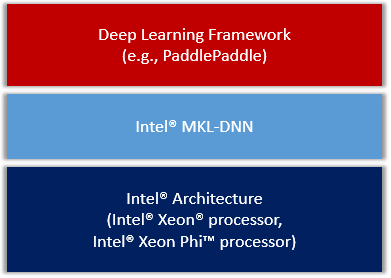
doc/design/mkl/image/overview.png
0 → 100644
10.5 KB
doc/design/mkl/mkl_packed.md
0 → 100644
doc/design/mkl/mkldnn.md
0 → 100644
doc/design/mkl/mkldnn_fluid.md
0 → 100644
doc/design/mkldnn/README.MD
已删除
100644 → 0
9.7 KB
doc/design/paddle_nccl.md
0 → 100644
doc/design/refactor/multi_cpu.md
0 → 100644
文件已添加
350.4 KB
76.3 KB
doc/design/support_new_device.md
0 → 100644
doc/design/switch_kernel.md
0 → 100644
43.4 KB
179.1 KB
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/howto/dev/write_docs_en.rst
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/howto/read_source.md
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
文件已移动
文件已移动
文件已移动
此差异已折叠。
文件已移动
文件已移动
文件已移动
此差异已折叠。
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/capi/error.cpp
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/framework/backward.md
已删除
100644 → 0
此差异已折叠。
paddle/framework/data_layout.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/framework/data_transform.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/framework/init.cc
0 → 100644
此差异已折叠。
paddle/framework/init.h
0 → 100644
此差异已折叠。
paddle/framework/init_test.cc
0 → 100644
此差异已折叠。
paddle/framework/library_type.h
0 → 100644
此差异已折叠。
paddle/framework/op_kernel_type.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/framework/tensor_util.cc
0 → 100644
此差异已折叠。
paddle/framework/tensor_util.cu
0 → 120000
此差异已折叠。
paddle/framework/tensor_util.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/framework/threadpool.cc
0 → 100644
此差异已折叠。
paddle/framework/threadpool.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/math/float16.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/math/tests/test_float16.cu
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/fill_op.cc
0 → 100644
此差异已折叠。
paddle/operators/ftrl_op.cc
0 → 100644
此差异已折叠。
paddle/operators/ftrl_op.cu
0 → 100644
此差异已折叠。
paddle/operators/ftrl_op.h
0 → 100644
此差异已折叠。
paddle/operators/hinge_loss_op.cc
0 → 100644
此差异已折叠。
paddle/operators/hinge_loss_op.cu
0 → 100644
此差异已折叠。
paddle/operators/hinge_loss_op.h
0 → 100644
此差异已折叠。
paddle/operators/log_loss_op.cc
0 → 100644
此差异已折叠。
paddle/operators/log_loss_op.cu
0 → 100644
此差异已折叠。
paddle/operators/log_loss_op.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/math/maxouting.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/math/unpooling.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/operators/maxout_op.cc
0 → 100644
此差异已折叠。
paddle/operators/maxout_op.cu.cc
0 → 100644
此差异已折叠。
paddle/operators/maxout_op.h
0 → 100644
此差异已折叠。
paddle/operators/nce_op.cc
0 → 100644
此差异已折叠。
paddle/operators/nce_op.h
0 → 100644
此差异已折叠。
文件已移动
此差异已折叠。
文件已移动
此差异已折叠。
文件已移动
paddle/operators/recv_op.cc
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/operators/reshape_op.cu
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/roi_pool_op.cc
0 → 100644
此差异已折叠。
paddle/operators/roi_pool_op.cu
0 → 100644
此差异已折叠。
paddle/operators/roi_pool_op.h
0 → 100644
此差异已折叠。
paddle/operators/row_conv_op.cc
0 → 100644
此差异已折叠。
paddle/operators/row_conv_op.cu
0 → 100644
此差异已折叠。
paddle/operators/row_conv_op.h
0 → 100644
此差异已折叠。
paddle/operators/send_op.cc
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/sequence_slice_op.cc
100755 → 100644
此差异已折叠。
paddle/operators/sequence_slice_op.h
100755 → 100644
此差异已折叠。
paddle/operators/spp_op.cc
0 → 100644
此差异已折叠。
paddle/operators/spp_op.cu.cc
0 → 100644
此差异已折叠。
paddle/operators/spp_op.h
0 → 100644
此差异已折叠。
paddle/operators/tensor.save
0 → 100644
此差异已折叠。
paddle/operators/unpool_op.cc
0 → 100644
此差异已折叠。
paddle/operators/unpool_op.cu.cc
0 → 100644
此差异已折叠。
paddle/operators/unpool_op.h
0 → 100644
此差异已折叠。
paddle/platform/cuda_profiler.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/platform/enforce.cc
0 → 100644
此差异已折叠。
paddle/platform/for_range.h
0 → 100644
此差异已折叠。
paddle/pybind/const_value.cc
0 → 100644
此差异已折叠。
paddle/pybind/const_value.h
0 → 100644
此差异已折叠。
paddle/scripts/check_env.sh
0 → 100755
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/trainer/tests/test.txt
已删除
100644 → 0
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
python/paddle/v2/fluid/clip.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。