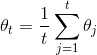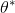Merge remote-tracking branch 'origin/develop' into feature/evaluator
Showing
doc/design/images/asgd.gif
0 → 100644
620 字节
doc/design/images/theta_star.gif
0 → 100644
156 字节
doc/design/parameter_average.md
0 → 100644