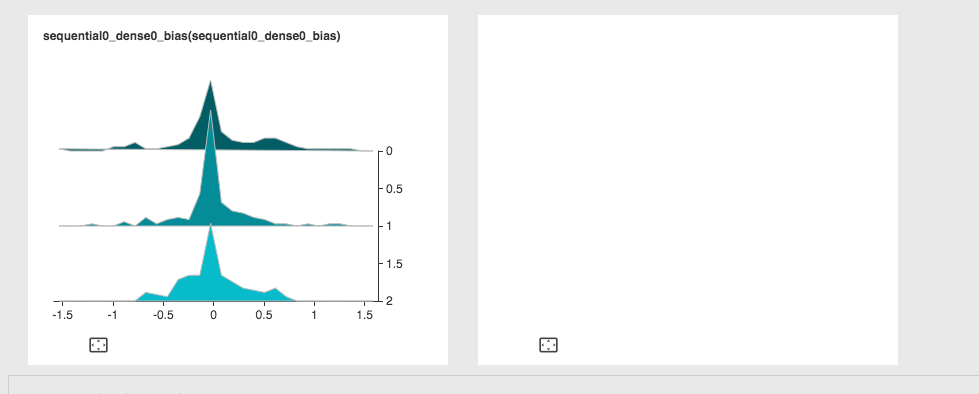
Cannot display more than one histogram at the same time
Created by: reminisce
I was trying VisualDL in an MXNet tutorial to track the gradients of two parameters for each epoch. However, only the histogram of the second gradient can be displayed. The changes I made to the tutorial are adding writer creation before the epoch loop:
param_grad_writers = {}
for name in param_names[:2]:
print(name)
with logger.mode(name):
param_grad_writers[name] = logger.histogram(name, num_buckets)and calling add_record at the end of each epoch loop:
grads = [i.grad() for i in net.collect_params().values()]
assert len(grads) == len(param_names)
for i, name in enumerate(param_names[:2]):
print('%d %s' % (i, name))
print(grads[i].shape)
assert name in param_grad_writers
assert np.prod(grads[i].shape) >= num_buckets
param_grad_writers[name].add_record(epoch, list(grads[i].asnumpy().flatten()))Here is the complete script I ran.
from __future__ import print_function
import random
from visualdl import LogWriter
import argparse
import logging
logging.basicConfig(level=logging.DEBUG)
import numpy as np
import mxnet as mx
from mxnet import gluon, autograd
from mxnet.gluon import nn
# Parse CLI arguments
parser = argparse.ArgumentParser(description='MXNet Gluon MNIST Example')
parser.add_argument('--batch-size', type=int, default=100,
help='batch size for training and testing (default: 100)')
parser.add_argument('--epochs', type=int, default=10,
help='number of epochs to train (default: 10)')
parser.add_argument('--lr', type=float, default=0.1,
help='learning rate (default: 0.1)')
parser.add_argument('--momentum', type=float, default=0.9,
help='SGD momentum (default: 0.9)')
parser.add_argument('--cuda', action='store_true', default=False,
help='Train on GPU with CUDA')
parser.add_argument('--log-interval', type=int, default=100, metavar='N',
help='how many batches to wait before logging training status')
opt = parser.parse_args()
logdir = "./tmp"
logger = LogWriter(logdir, sync_cycle=10)
# mark the components with 'train' label.
with logger.mode("train"):
train_acc = logger.scalar("scalars/accuracy")
with logger.mode('valid'):
valid_acc = logger.scalar("scalars/accuracy")
# define network
net = nn.Sequential()
with net.name_scope():
net.add(nn.Dense(128, activation='relu'))
net.add(nn.Dense(64, activation='relu'))
net.add(nn.Dense(10))
# data
def transformer(data, label):
data = data.reshape((-1,)).astype(np.float32)/255
return data, label
train_data = gluon.data.DataLoader(
gluon.data.vision.MNIST('./data', train=True, transform=transformer),
batch_size=opt.batch_size, shuffle=True, last_batch='discard')
val_data = gluon.data.DataLoader(
gluon.data.vision.MNIST('./data', train=False, transform=transformer),
batch_size=opt.batch_size, shuffle=False)
# train
def test(ctx):
metric = mx.metric.Accuracy()
for data, label in val_data:
data = data.as_in_context(ctx)
label = label.as_in_context(ctx)
output = net(data)
metric.update([label], [output])
return metric.get()
def train(epochs, ctx):
# Collect all parameters from net and its children, then initialize them.
net.initialize(mx.init.Xavier(magnitude=2.24), ctx=ctx)
# Trainer is for updating parameters with gradient.
trainer = gluon.Trainer(net.collect_params(), 'sgd',
{'learning_rate': opt.lr, 'momentum': opt.momentum})
metric = mx.metric.Accuracy()
loss = gluon.loss.SoftmaxCrossEntropyLoss()
params = net.collect_params()
param_names = params.keys()
print(param_names)
num_buckets = 100
param_grad_writers = {}
for name in param_names[:2]:
print(name)
with logger.mode(name):
param_grad_writers[name] = logger.histogram(name, num_buckets)
for epoch in range(epochs):
# reset data iterator and metric at begining of epoch.
metric.reset()
for i, (data, label) in enumerate(train_data):
# Copy data to ctx if necessary
data = data.as_in_context(ctx)
label = label.as_in_context(ctx)
# Start recording computation graph with record() section.
# Recorded graphs can then be differentiated with backward.
with autograd.record():
output = net(data)
L = loss(output, label)
L.backward()
# take a gradient step with batch_size equal to data.shape[0]
trainer.step(data.shape[0])
# update metric at last.
metric.update([label], [output])
if i % opt.log_interval == 0 and i > 0:
name, acc = metric.get()
print('[Epoch %d Batch %d] Training: %s=%f'%(epoch, i, name, acc))
grads = [i.grad() for i in net.collect_params().values()]
assert len(grads) == len(param_names)
for i, name in enumerate(param_names[:2]):
print('%d %s' % (i, name))
print(grads[i].shape)
assert name in param_grad_writers
assert np.prod(grads[i].shape) >= num_buckets
param_grad_writers[name].add_record(epoch, list(grads[i].asnumpy().flatten()))
name, acc = metric.get()
print('[Epoch %d] Training: %s=%f'%(epoch, name, acc))
name, val_acc = test(ctx)
print('[Epoch %d] Validation: %s=%f'%(epoch, name, val_acc))
train_acc.add_record(epoch, acc)
valid_acc.add_record(epoch, val_acc)
net.save_params('mnist.params')
if __name__ == '__main__':
if opt.cuda:
ctx = mx.gpu(0)
else:
ctx = mx.cpu()
train(opt.epochs, ctx)