Merge pull request #8 from PaddlePaddle/develop
00
Showing
docs/appendix/interpret.md
0 → 100644
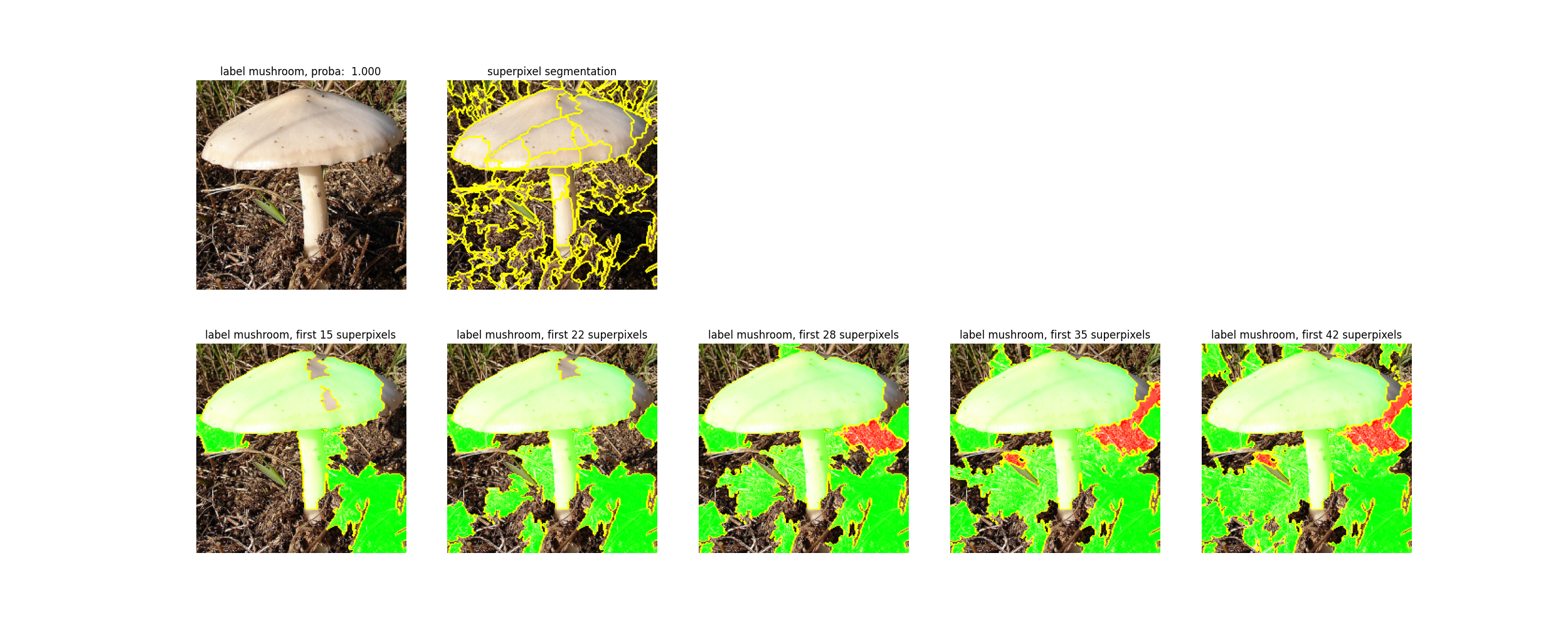
docs/images/lime.png
0 → 100644
802.8 KB
docs/images/normlime.png
0 → 100644
1.5 MB
tools/codestyle/clang_format.hook
0 → 100755