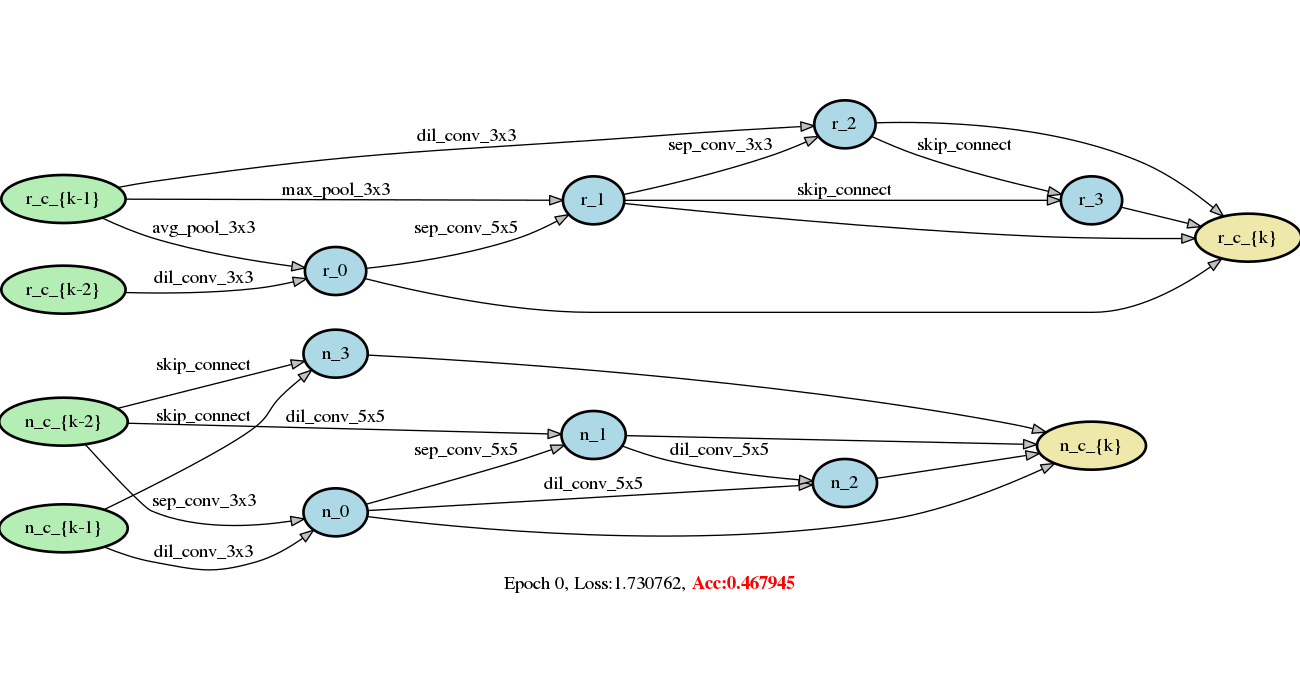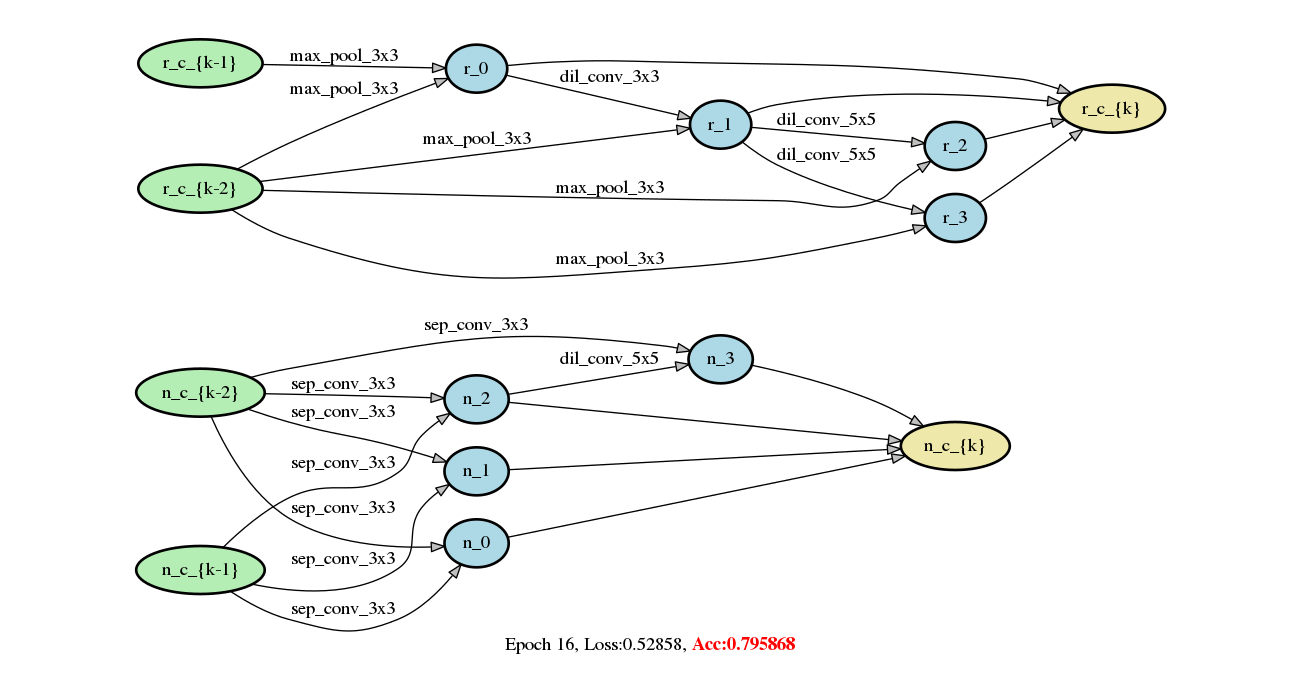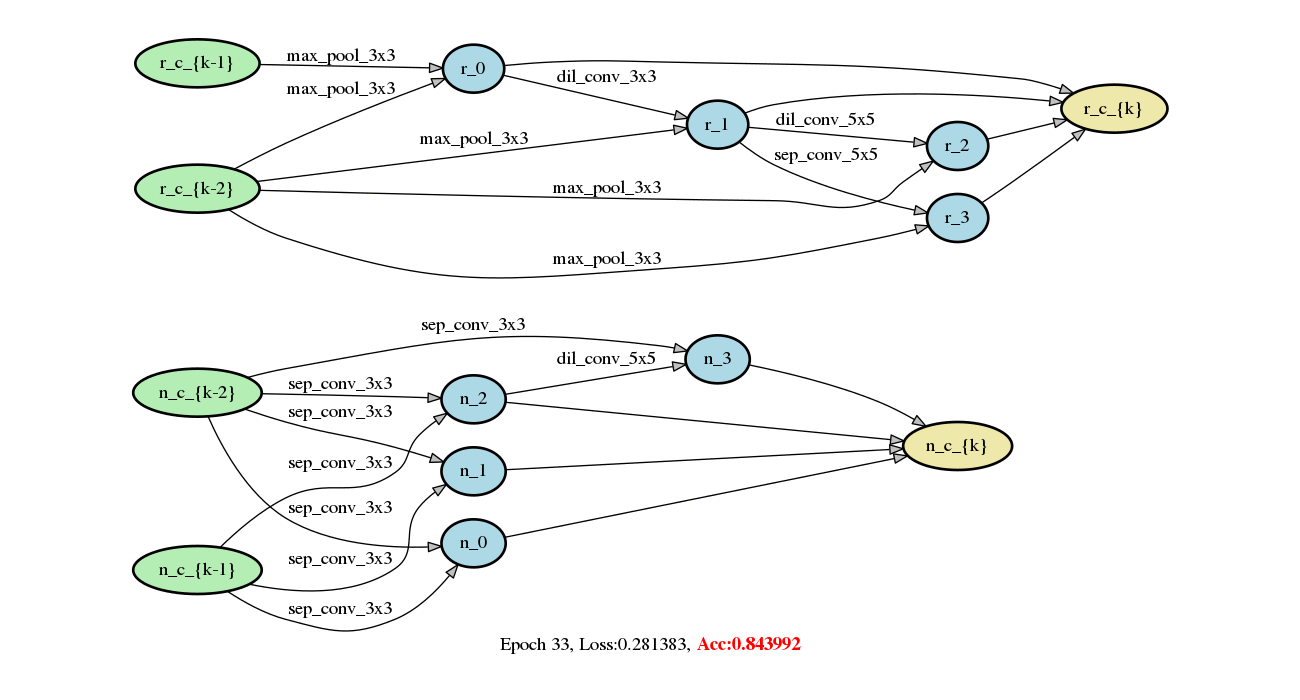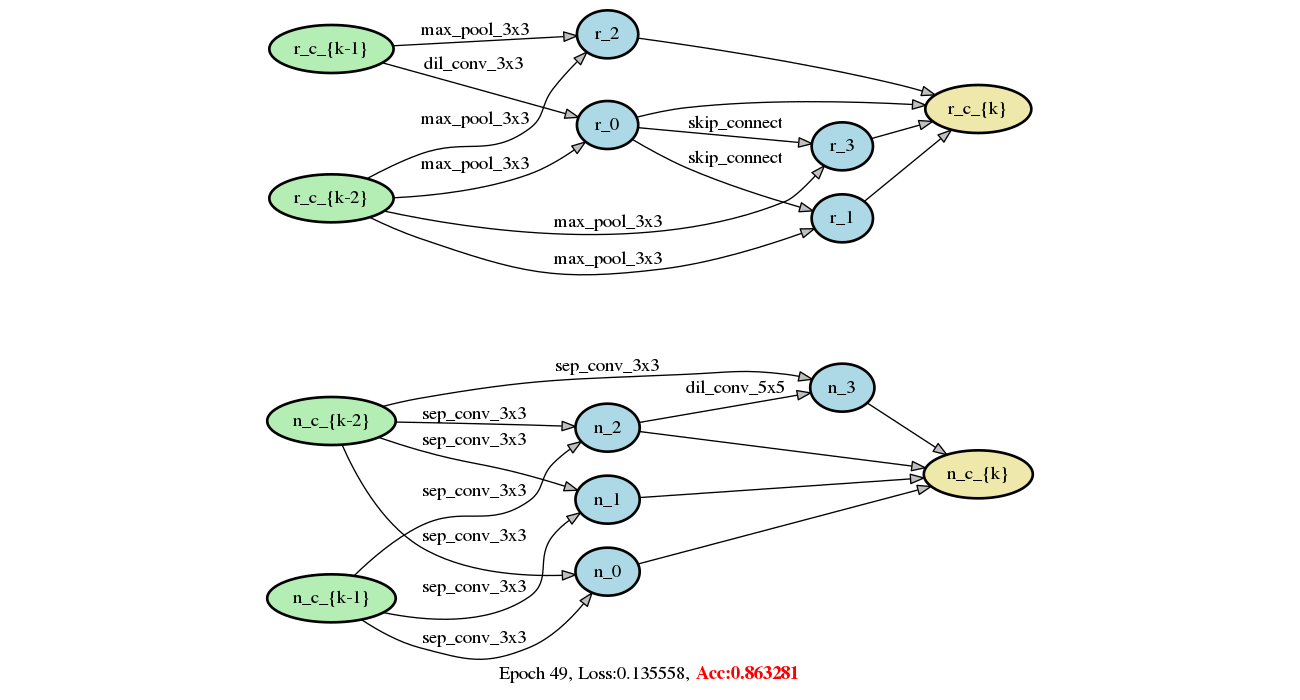
Add DARTS series methods (#213)
Showing
demo/darts/README.md
0 → 100644
demo/darts/genotypes.py
0 → 100644
demo/darts/images/networks.gif
0 → 100755
2.2 MB
demo/darts/model.py
0 → 100644
demo/darts/model_search.py
0 → 100644
demo/darts/operations.py
0 → 100644
demo/darts/reader.py
0 → 100644
demo/darts/search.py
0 → 100644
demo/darts/train.py
0 → 100644
demo/darts/train_imagenet.py
0 → 100644
demo/darts/visualize.py
0 → 100644
paddleslim/common/meter.py
0 → 100644
paddleslim/nas/darts/__init__.py
0 → 100644
paddleslim/nas/darts/architect.py
0 → 100644