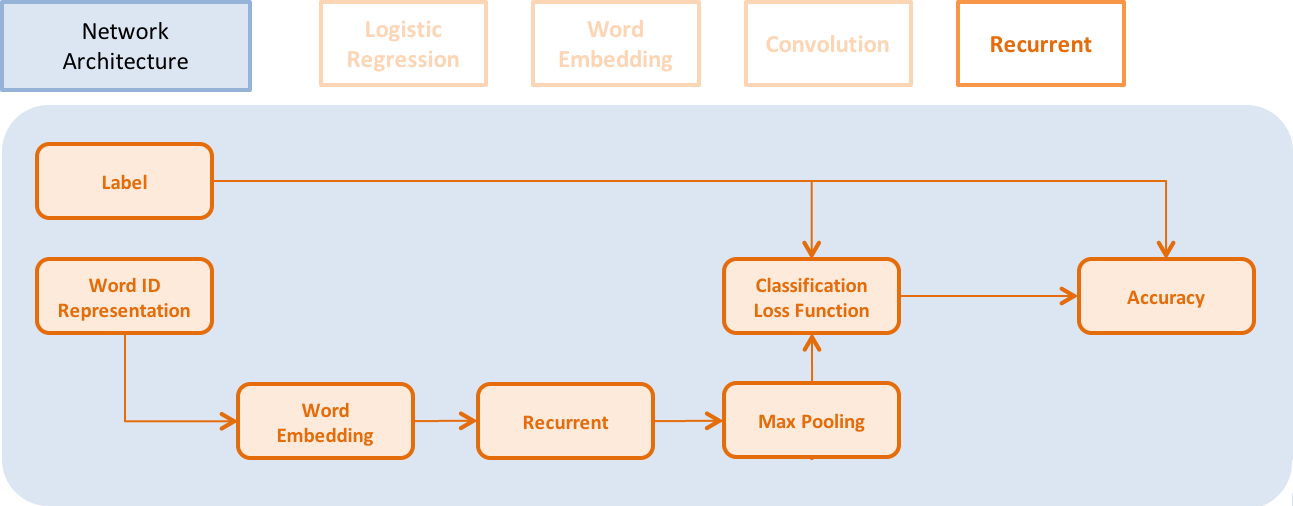
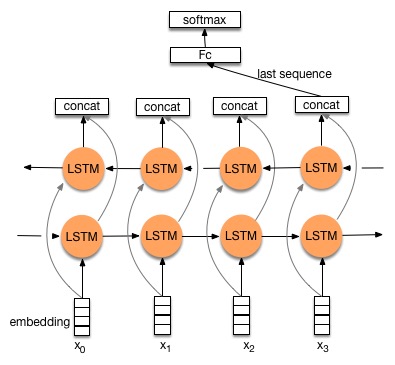
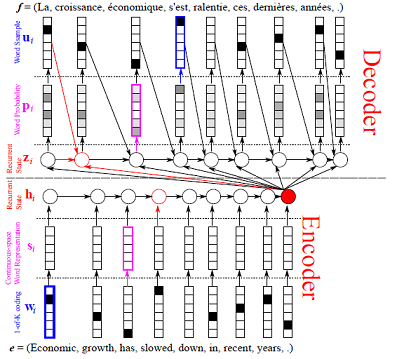
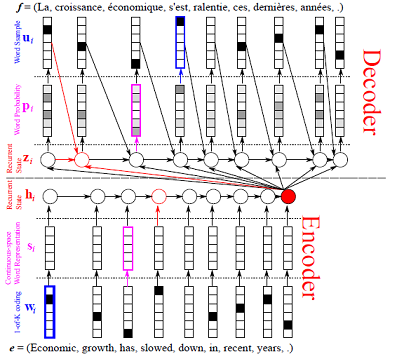
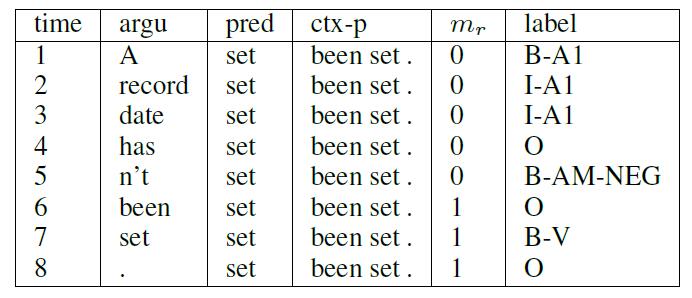
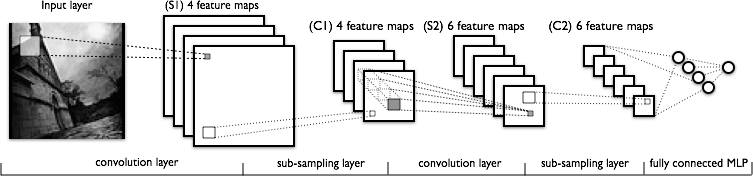
Update doc
Showing
doc/html/.buildinfo
0 → 100644
49.7 KB
59.0 KB
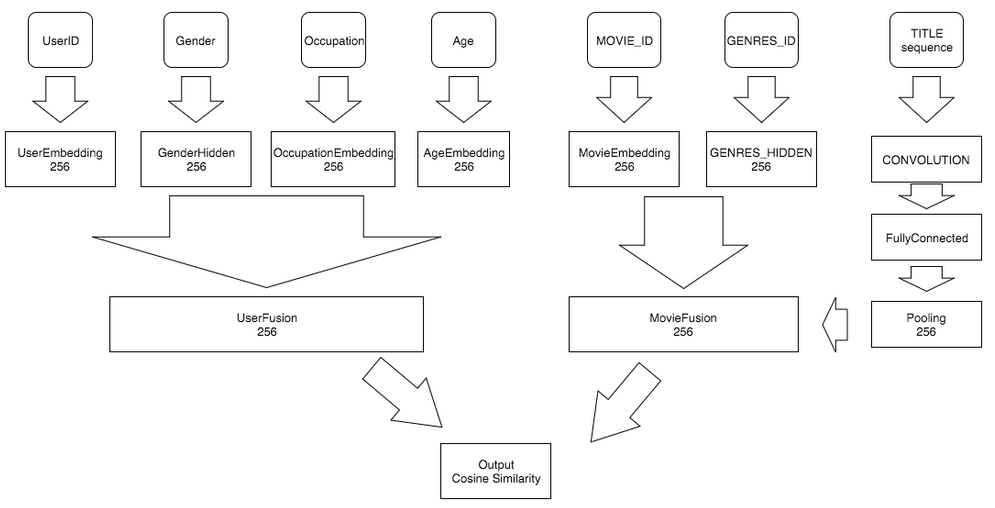
doc/html/_images/NetConv_en.png
0 → 100644
67.3 KB
doc/html/_images/NetLR_en.png
0 → 100644
49.8 KB
doc/html/_images/NetRNN_en.png
0 → 100644
57.1 KB
7.3 KB
13.8 KB
13.8 KB
doc/html/_images/Pipeline_en.jpg
0 → 100644
11.4 KB
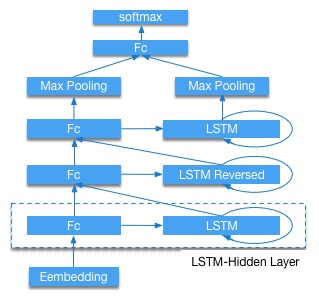
doc/html/_images/bi_lstm.jpg
0 → 100644
34.8 KB
doc/html/_images/bi_lstm1.jpg
0 → 100644
34.8 KB
doc/html/_images/cifar.png
0 → 100644
455.6 KB
66.5 KB
66.5 KB
doc/html/_images/feature.jpg
0 → 100644
30.5 KB
51.4 KB
doc/html/_images/lenet.png
0 → 100644
48.7 KB
doc/html/_images/lstm.png
0 → 100644
49.5 KB
doc/html/_images/network_arch.png
0 → 100644
27.2 KB
66.9 KB
doc/html/_images/plot.png
0 → 100644
30.3 KB
81.2 KB
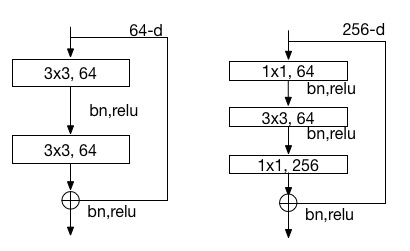
doc/html/_images/resnet_block.jpg
0 → 100644
21.9 KB
doc/html/_images/stacked_lstm.jpg
0 → 100644
30.3 KB
doc/html/_sources/build/index.txt
0 → 100644
doc/html/_sources/demo/index.txt
0 → 100644
此差异已折叠。
doc/html/_sources/index.txt
0 → 100644
doc/html/_sources/layer.txt
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/_sources/ui/index.txt
0 → 100644
此差异已折叠。
此差异已折叠。
doc/html/_static/ajax-loader.gif
0 → 100644
此差异已折叠。
doc/html/_static/basic.css
0 → 100644
此差异已折叠。
doc/html/_static/classic.css
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/_static/comment.png
0 → 100644
此差异已折叠。
doc/html/_static/doctools.js
0 → 100644
此差异已折叠。
doc/html/_static/down-pressed.png
0 → 100644
此差异已折叠。
doc/html/_static/down.png
0 → 100644
此差异已折叠。
doc/html/_static/file.png
0 → 100644
此差异已折叠。
doc/html/_static/jquery-1.11.1.js
0 → 100644
此差异已折叠。
doc/html/_static/jquery.js
0 → 100644
此差异已折叠。
doc/html/_static/minus.png
0 → 100644
此差异已折叠。
doc/html/_static/plus.png
0 → 100644
此差异已折叠。
doc/html/_static/pygments.css
0 → 100644
此差异已折叠。
doc/html/_static/searchtools.js
0 → 100644
此差异已折叠。
doc/html/_static/sidebar.js
0 → 100644
此差异已折叠。
此差异已折叠。
doc/html/_static/underscore.js
0 → 100644
此差异已折叠。
doc/html/_static/up-pressed.png
0 → 100644
此差异已折叠。
doc/html/_static/up.png
0 → 100644
此差异已折叠。
doc/html/_static/websupport.js
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/build/index.html
0 → 100644
此差异已折叠。
doc/html/cluster/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/demo/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/demo/rec/ml_dataset.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/genindex.html
0 → 100644
此差异已折叠。
doc/html/index.html
0 → 100644
此差异已折叠。
doc/html/layer.html
0 → 100644
此差异已折叠。
doc/html/objects.inv
0 → 100644
此差异已折叠。
doc/html/py-modindex.html
0 → 100644
此差异已折叠。
doc/html/search.html
0 → 100644
此差异已折叠。
doc/html/searchindex.js
0 → 100644
此差异已折叠。
doc/html/source/api/api.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/source/cuda/rnn/rnn.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/source/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/source/utils/enum.html
0 → 100644
此差异已折叠。
doc/html/source/utils/lock.html
0 → 100644
此差异已折叠。
doc/html/source/utils/queue.html
0 → 100644
此差异已折叠。
doc/html/source/utils/thread.html
0 → 100644
此差异已折叠。
此差异已折叠。
doc/html/ui/api/rnn/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc/html/ui/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/.buildinfo
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_images/NetConv.jpg
0 → 100644
此差异已折叠。
doc_cn/html/_images/NetLR.jpg
0 → 100644
此差异已折叠。
doc_cn/html/_images/NetRNN.jpg
0 → 100644
此差异已折叠。
doc_cn/html/_images/Pipeline.jpg
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/_sources/index.txt
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/_sources/ui/index.txt
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/basic.css
0 → 100644
此差异已折叠。
doc_cn/html/_static/classic.css
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/comment.png
0 → 100644
此差异已折叠。
doc_cn/html/_static/doctools.js
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/down.png
0 → 100644
此差异已折叠。
doc_cn/html/_static/file.png
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/jquery.js
0 → 100644
此差异已折叠。
doc_cn/html/_static/minus.png
0 → 100644
此差异已折叠。
doc_cn/html/_static/plus.png
0 → 100644
此差异已折叠。
doc_cn/html/_static/pygments.css
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/sidebar.js
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/underscore.js
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/_static/up.png
0 → 100644
此差异已折叠。
doc_cn/html/_static/websupport.js
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/cluster/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/demo/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/genindex.html
0 → 100644
此差异已折叠。
doc_cn/html/index.html
0 → 100644
此差异已折叠。
doc_cn/html/objects.inv
0 → 100644
此差异已折叠。
doc_cn/html/search.html
0 → 100644
此差异已折叠。
doc_cn/html/searchindex.js
0 → 100644
此差异已折叠。
此差异已折叠。
doc_cn/html/ui/cmd/index.html
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
doc_cn/html/ui/index.html
0 → 100644
此差异已折叠。
此差异已折叠。