Merge pull request #233 from tianxin1860/develop
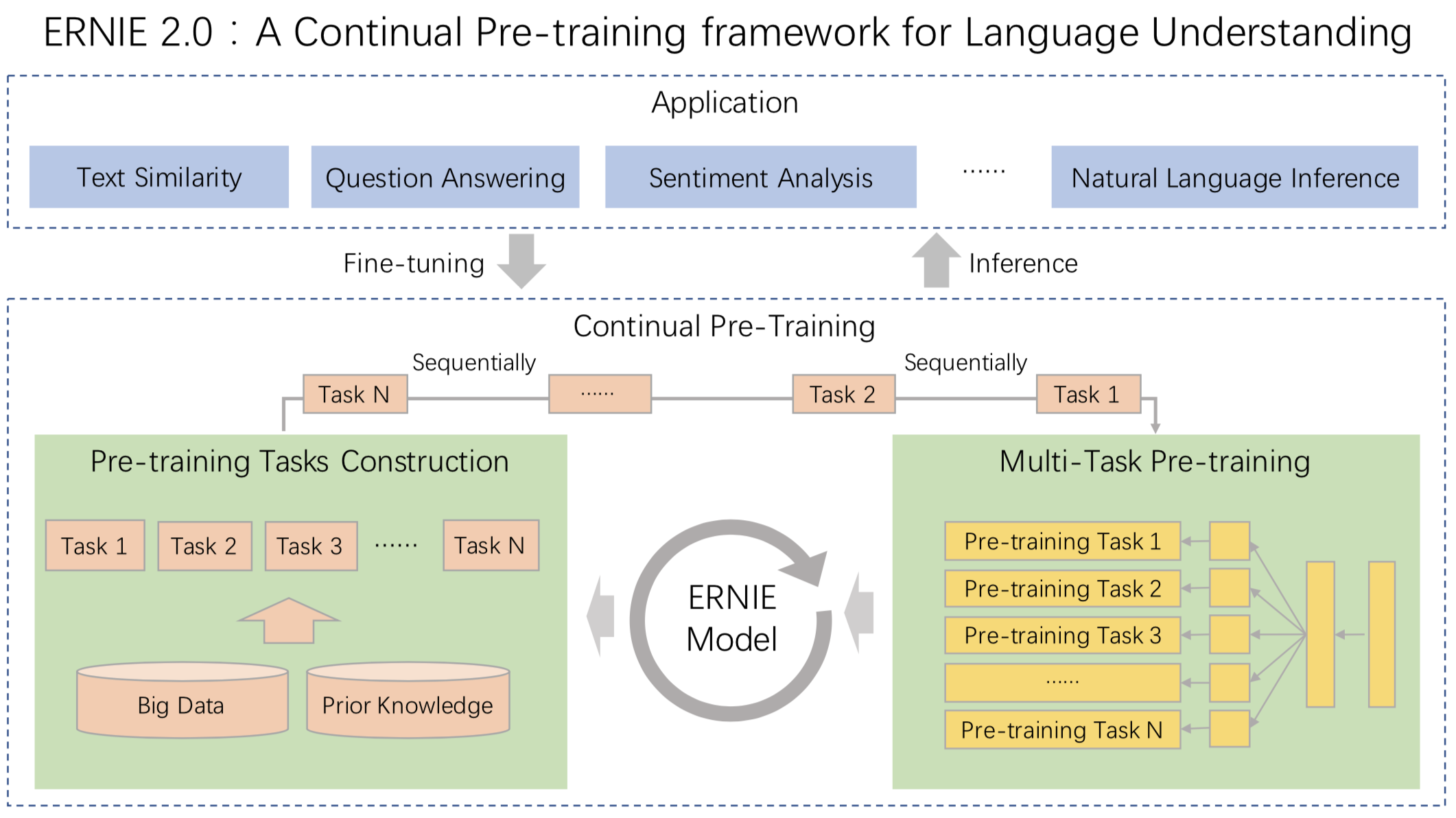
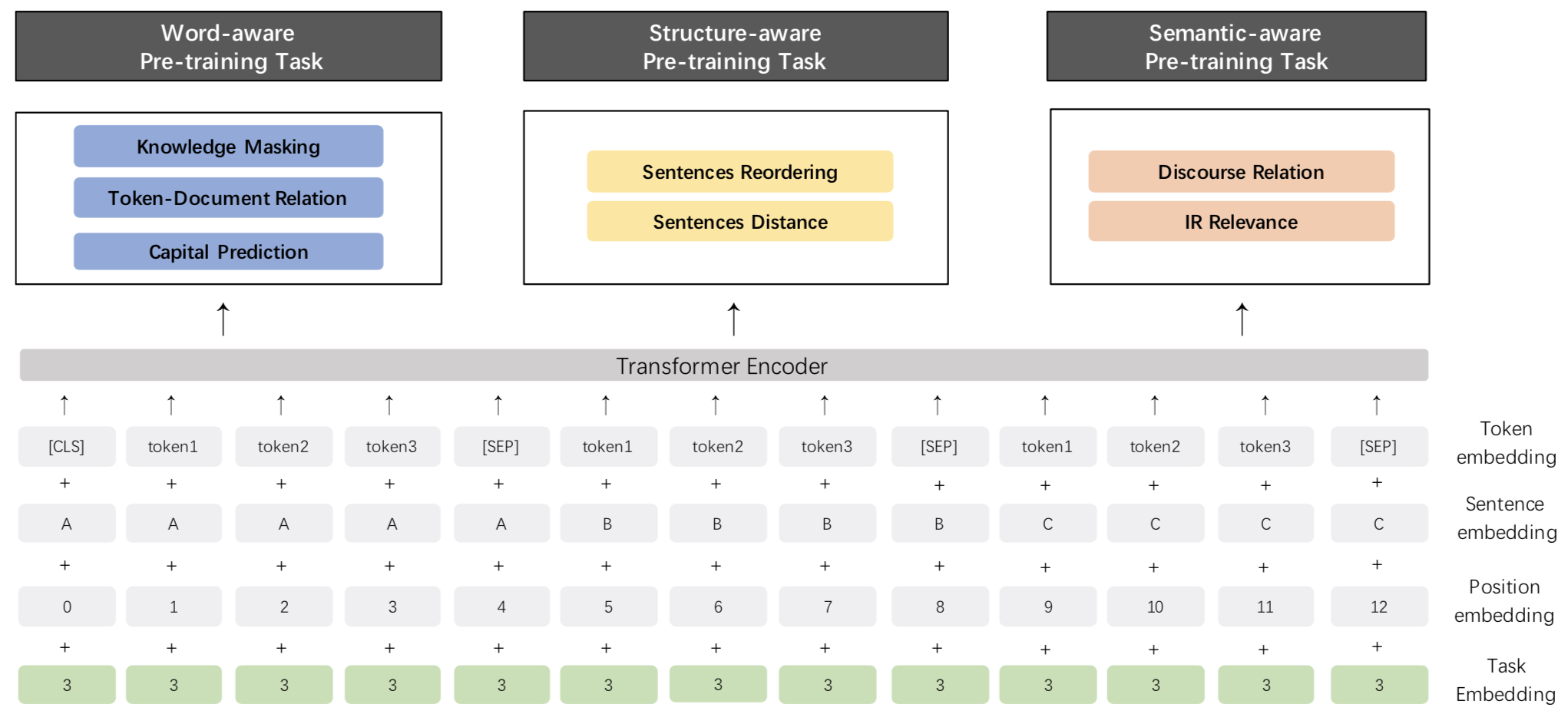
Release ERNIE 2.0
Showing
.metas/ernie2.0_arch.png
0 → 100644
303.4 KB
.metas/ernie2.0_model.png
0 → 100644
169.8 KB
此差异已折叠。
README.zh.md
0 → 100644
此差异已折叠。
classify_infer.py
0 → 100644
config/vocab_en.txt
0 → 100644
此差异已折叠。
finetune/mrc.py
0 → 100644
model/ernie.py
0 → 100644
run_classifier.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
script/en_glue/preprocess/cvt.sh
0 → 100644
此差异已折叠。
script/en_glue/preprocess/mnli.py
0 → 100644
此差异已折叠。
script/en_glue/preprocess/qnli.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
utils/cmrc2018_eval.py
0 → 100644
此差异已折叠。