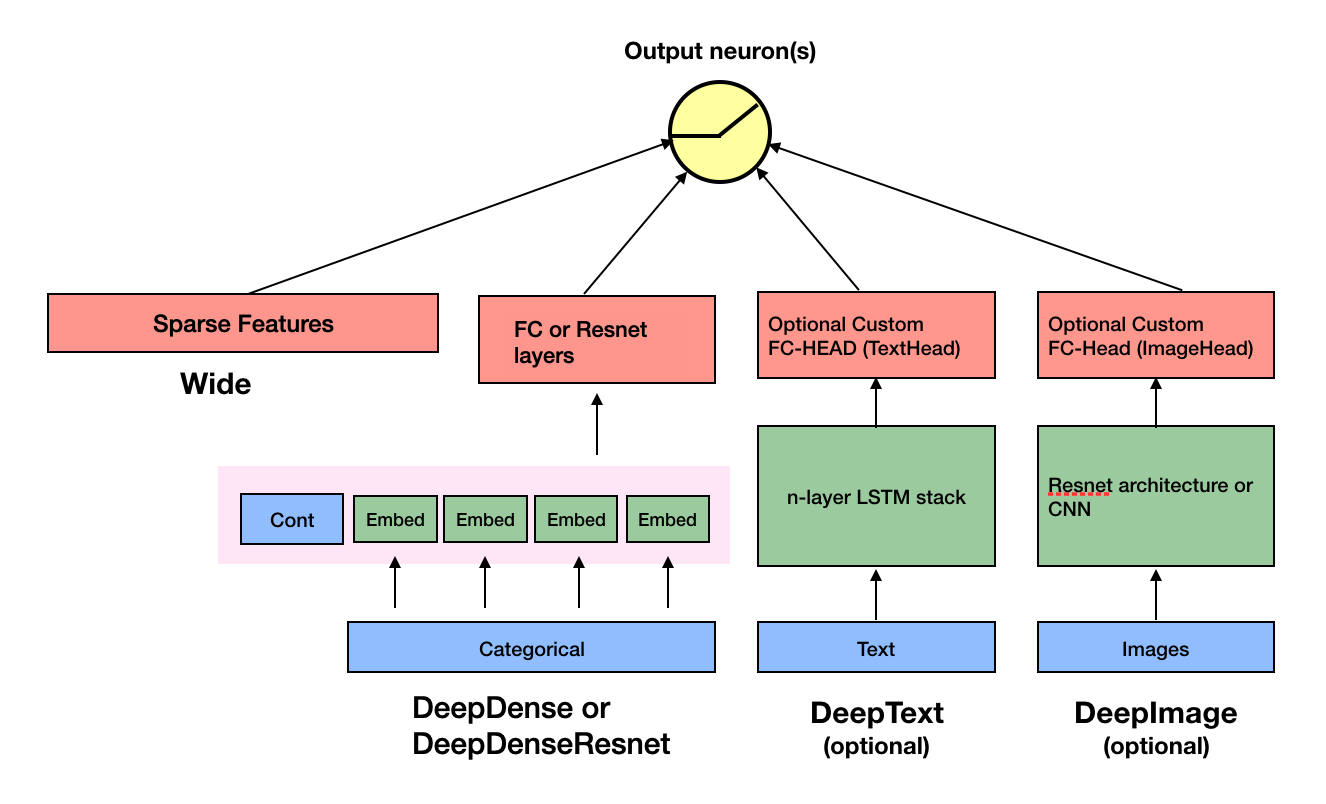
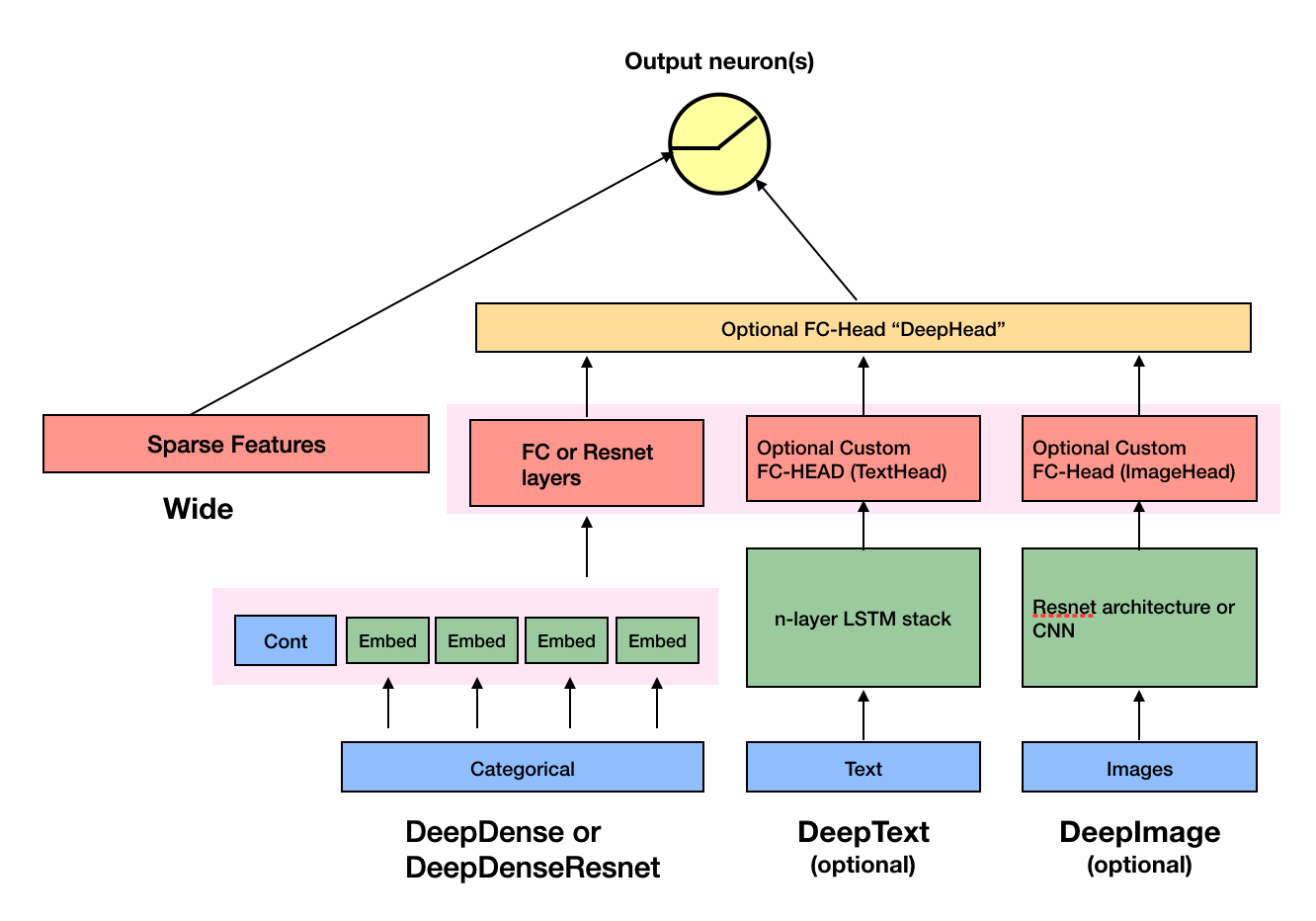
changed the dense layer to be almost identical to that of fastai, which I...
changed the dense layer to be almost identical to that of fastai, which I really like. Changed the code accordingly. Changed the name of DeepDense and DeepDenseResnet to TabMlp and TabResnet. Change the tests acccordingly
Showing
116.7 KB
126.4 KB
72.8 KB
38.0 KB
86.5 KB
文件已移动