update code
Showing
15.1 KB
15.6 KB
14.1 KB
| W: | H:
| W: | H:


13.7 KB
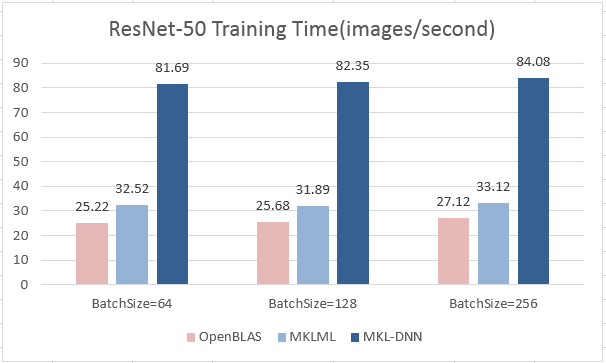
| W: | H:
| W: | H:


benchmark/figs/vgg-cpu-infer.png
0 → 100644
13.7 KB
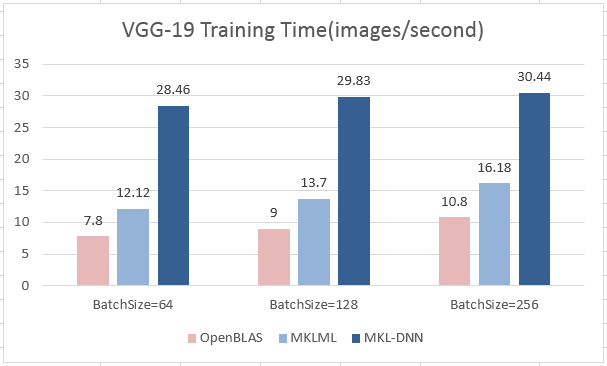
| W: | H:
| W: | H:


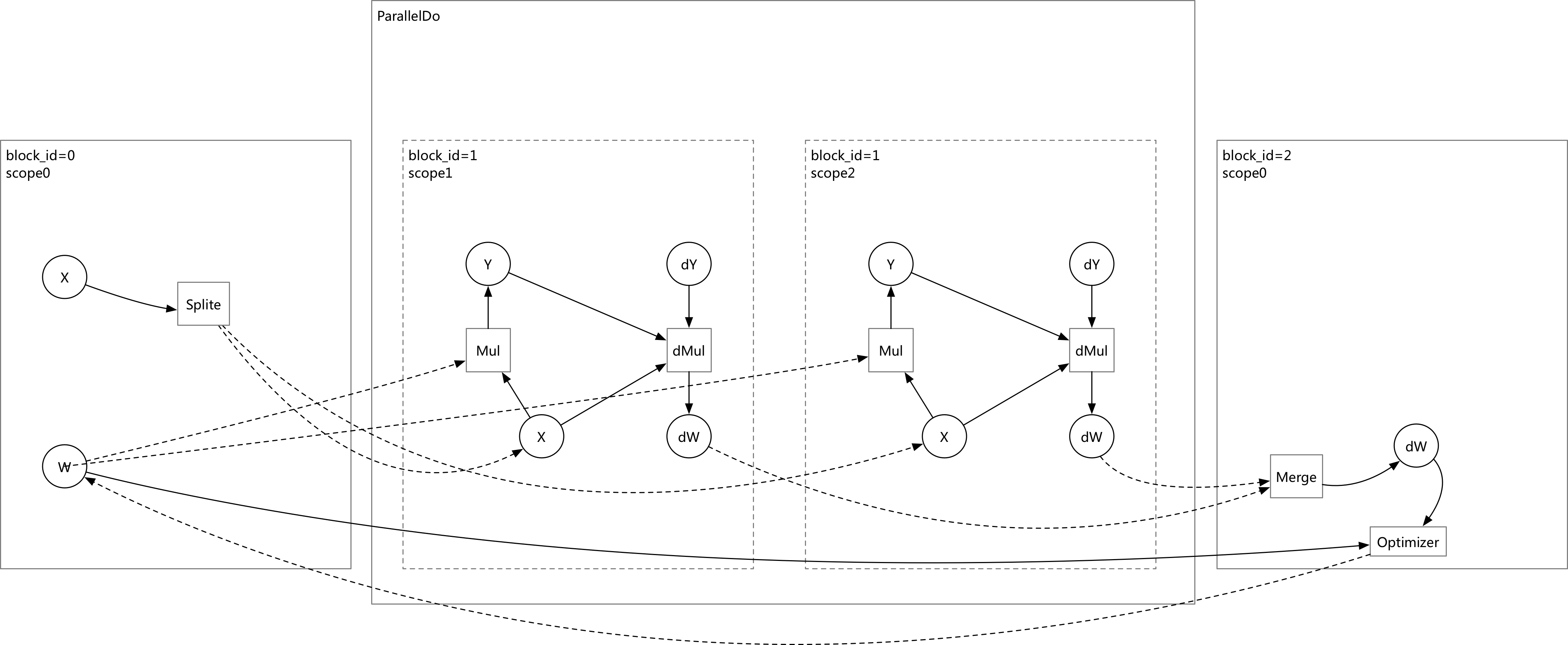
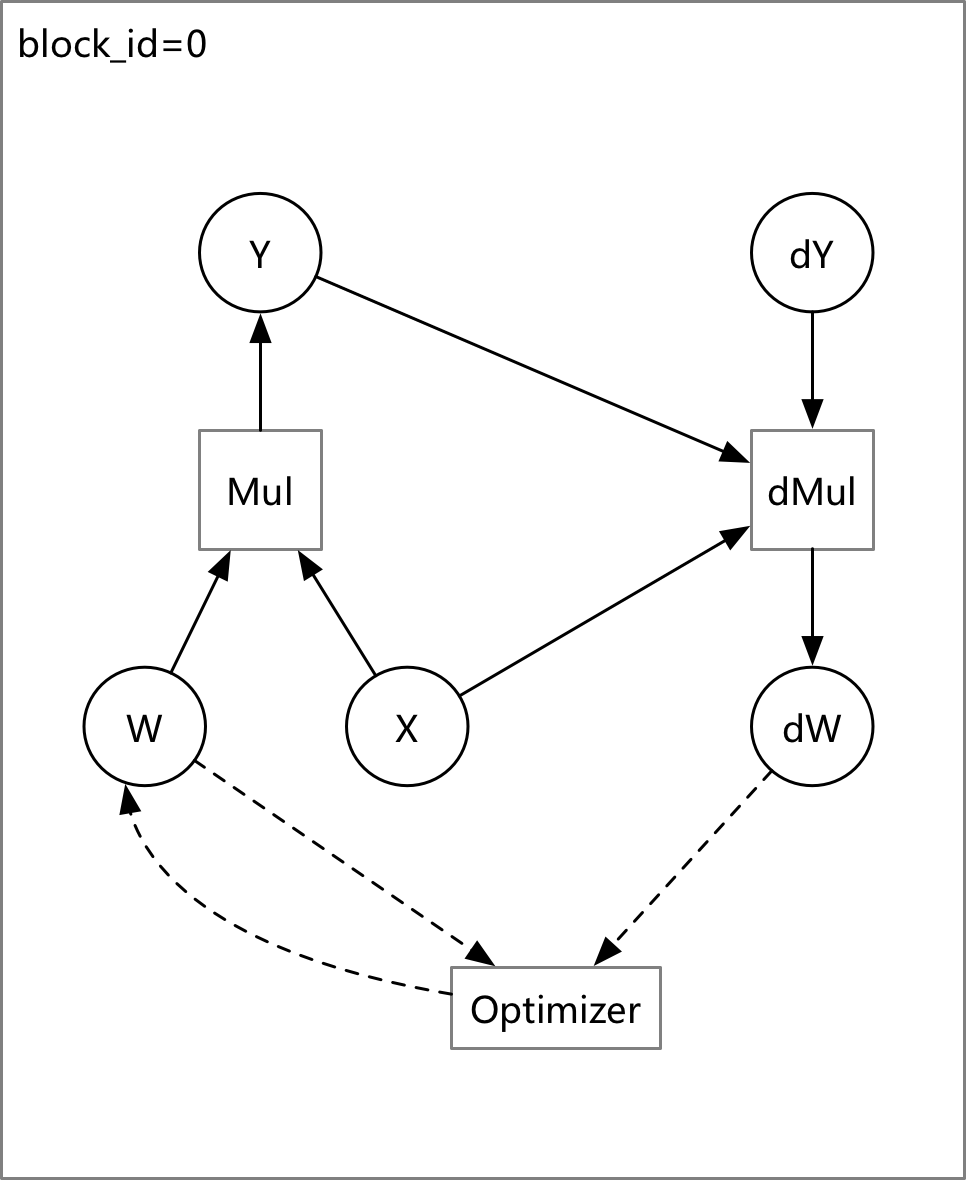
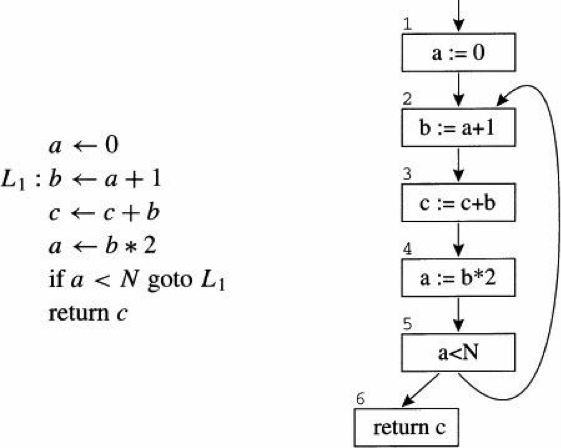
doc/design/backward.md
0 → 100644
doc/design/ci_build_whl.png
0 → 100644
280.4 KB
文件已移动
文件已移动
文件已移动
文件已移动
189.2 KB
文件已移动
文件已移动
文件已添加
102.5 KB
文件已添加
350.4 KB
76.3 KB
文件已移动
文件已移动
文件已添加
134.5 KB
doc/design/error_clip.md
0 → 100644
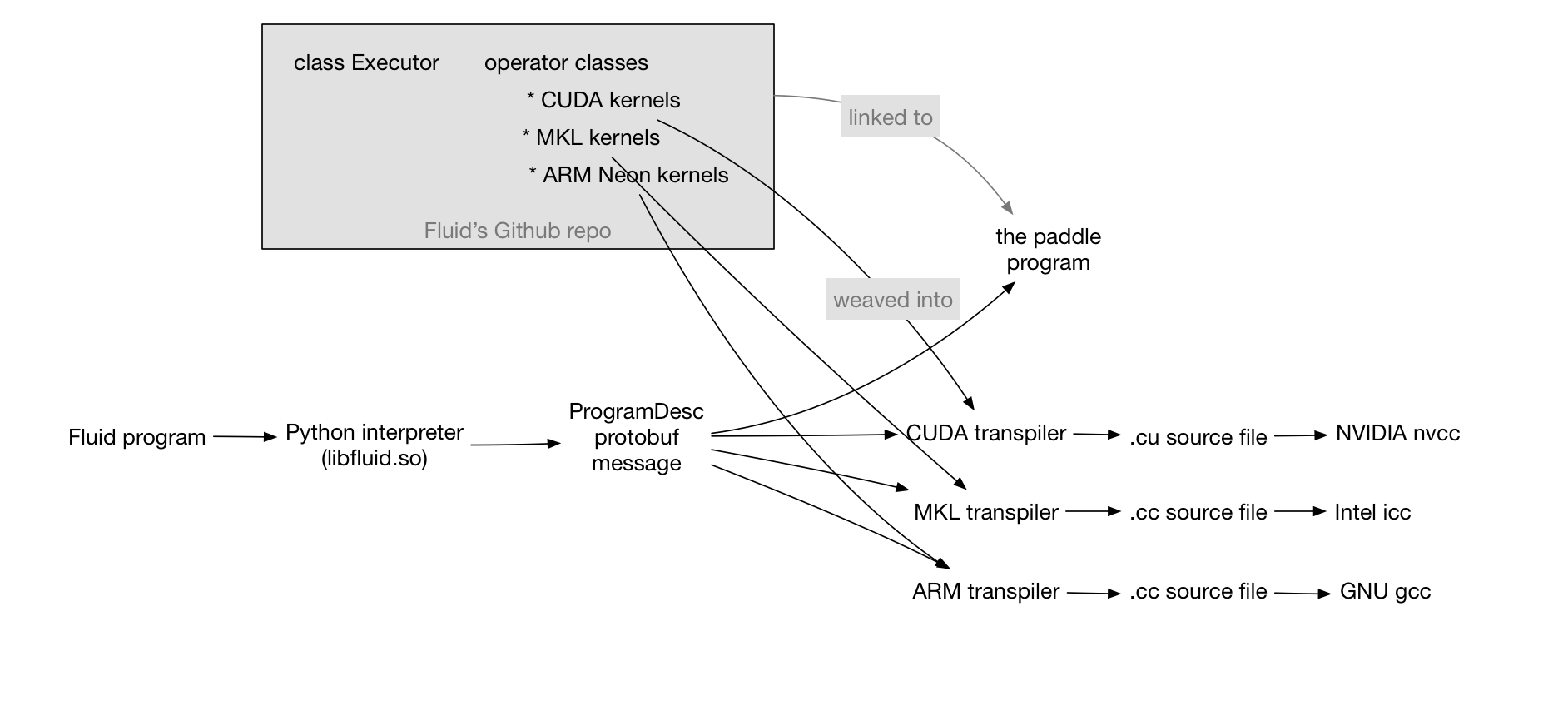
doc/design/fluid-compiler.graffle
0 → 100644
文件已添加
doc/design/fluid-compiler.png
0 → 100644
121.2 KB
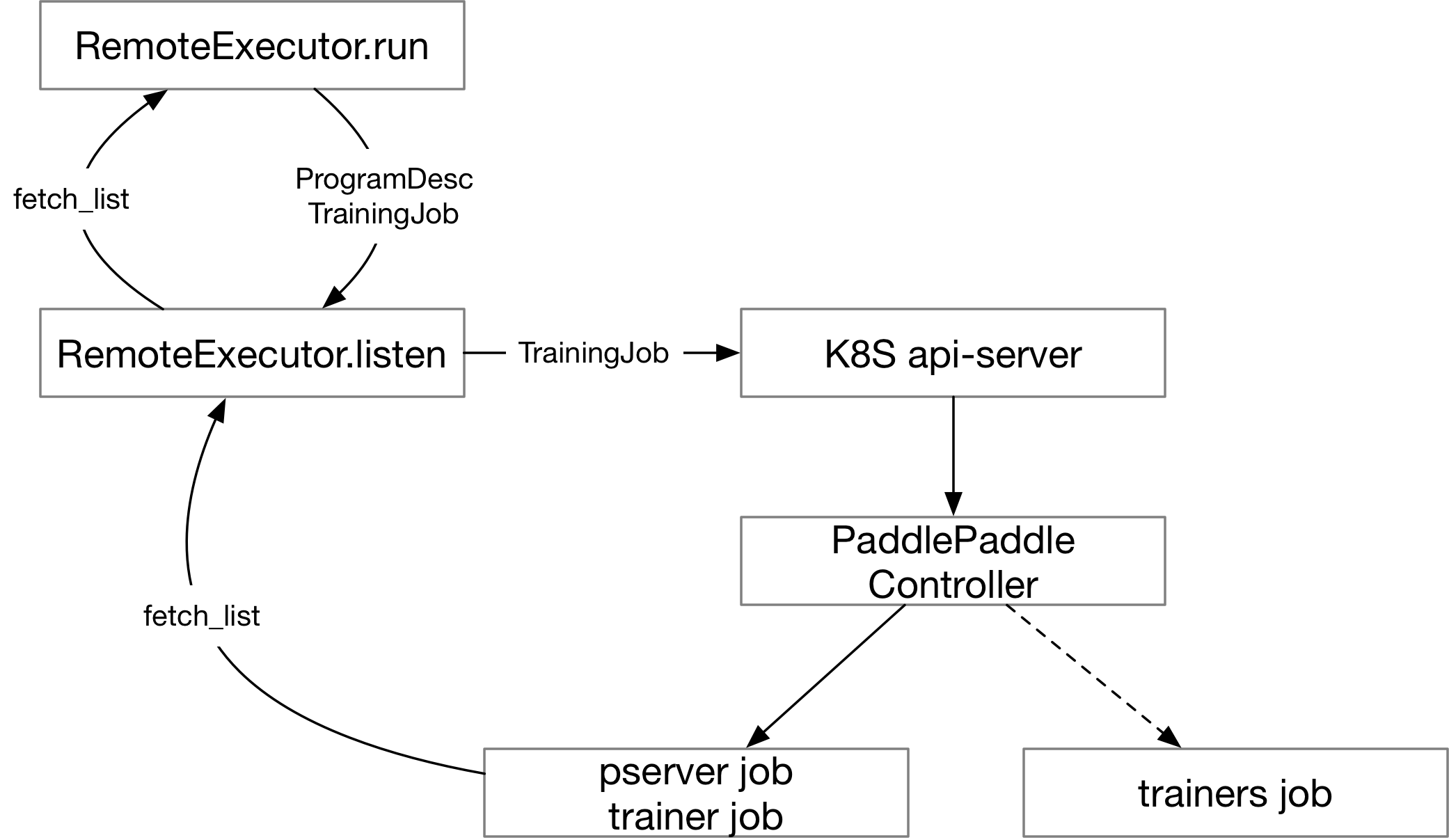
doc/design/fluid.md
0 → 100644
83.3 KB
22.5 KB
39.7 KB
文件已移动
文件已移动
文件已移动
文件已添加
108.4 KB
文件已添加
32.8 KB
doc/design/images/profiler.png
0 → 100644
49.9 KB
doc/design/kernel_hint_design.md
0 → 100644
doc/design/memory_optimization.md
0 → 100644
doc/design/mkl/mkl_packed.md
0 → 100644
doc/design/mkl/mkldnn_fluid.md
0 → 100644
doc/design/paddle_nccl.md
0 → 100644
doc/design/profiler.md
0 → 100644
46.5 KB
文件已删除
28.3 KB
doc/design/support_new_device.md
0 → 100644
doc/design/switch_kernel.md
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
文件已移动
文件已移动
文件已移动
此差异已折叠。
文件已移动
文件已移动
文件已移动
此差异已折叠。
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
文件已移动
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/framework/backward.md
已删除
100644 → 0
此差异已折叠。
paddle/framework/data_layout.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/framework/data_transform.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/framework/init.cc
0 → 100644
此差异已折叠。
paddle/framework/init.h
0 → 100644
此差异已折叠。
paddle/framework/init_test.cc
0 → 100644
此差异已折叠。
paddle/framework/library_type.h
0 → 100644
此差异已折叠。
paddle/framework/op_kernel_type.h
0 → 100644
此差异已折叠。
此差异已折叠。
paddle/framework/tensor_util.cc
0 → 100644
此差异已折叠。
paddle/framework/tensor_util.cu
0 → 120000
此差异已折叠。
此差异已折叠。
paddle/framework/threadpool.cc
0 → 100644
此差异已折叠。
paddle/framework/threadpool.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/inference/CMakeLists.txt
0 → 100644
此差异已折叠。
paddle/inference/example.cc
0 → 100644
此差异已折叠。
paddle/inference/inference.cc
0 → 100644
此差异已折叠。
paddle/inference/inference.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/get_places_op.cc
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/norm_op.cc
0 → 100644
此差异已折叠。
paddle/operators/norm_op.cu
0 → 100644
此差异已折叠。
paddle/operators/norm_op.h
0 → 100644
此差异已折叠。
文件已移动
此差异已折叠。
文件已移动
此差异已折叠。
文件已移动
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/operators/spp_op.cc
0 → 100644
此差异已折叠。
paddle/operators/spp_op.cu.cc
0 → 100644
此差异已折叠。
paddle/operators/spp_op.h
0 → 100644
此差异已折叠。
paddle/operators/tensor.save
已删除
100644 → 0
此差异已折叠。
paddle/operators/warpctc_op.cc
0 → 100644
此差异已折叠。
paddle/operators/warpctc_op.cu.cc
0 → 100644
此差异已折叠。
paddle/operators/warpctc_op.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
paddle/platform/dynload/warpctc.h
0 → 100644
此差异已折叠。
paddle/platform/for_range.h
0 → 100644
此差异已折叠。
paddle/platform/mkldnn_helper.h
0 → 100644
此差异已折叠。
paddle/platform/profiler.cc
0 → 100644
此差异已折叠。
paddle/platform/profiler.h
0 → 100644
此差异已折叠。
paddle/platform/profiler_test.cc
0 → 100644
此差异已折叠。
paddle/pybind/const_value.cc
0 → 100644
此差异已折叠。
paddle/pybind/const_value.h
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
python/paddle/v2/fluid/clip.py
0 → 100644
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。
此差异已折叠。