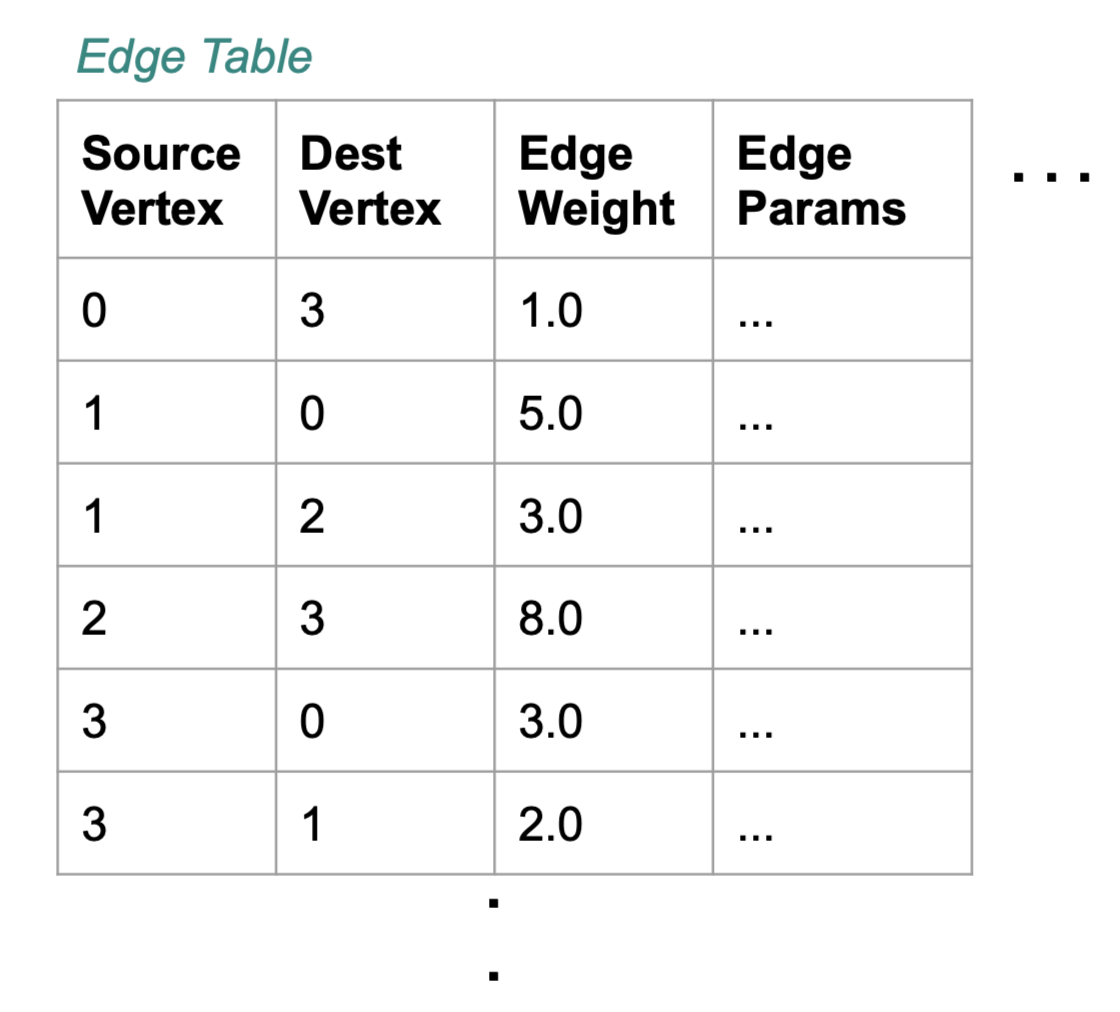
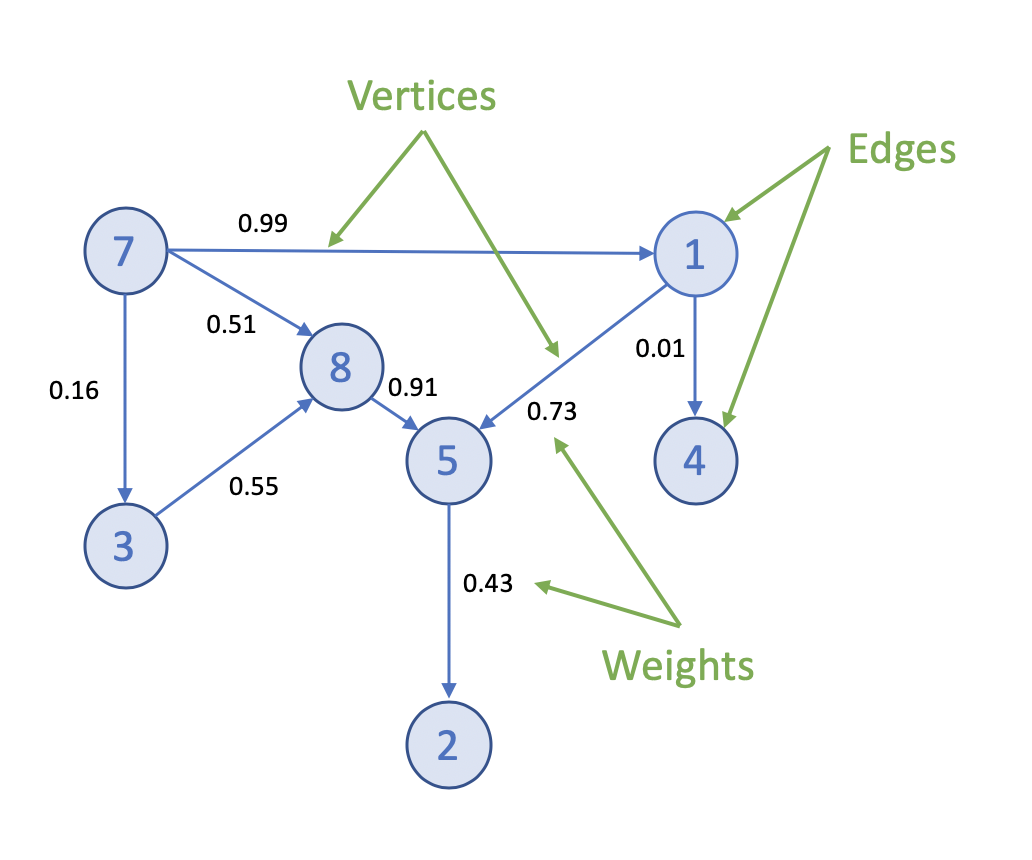
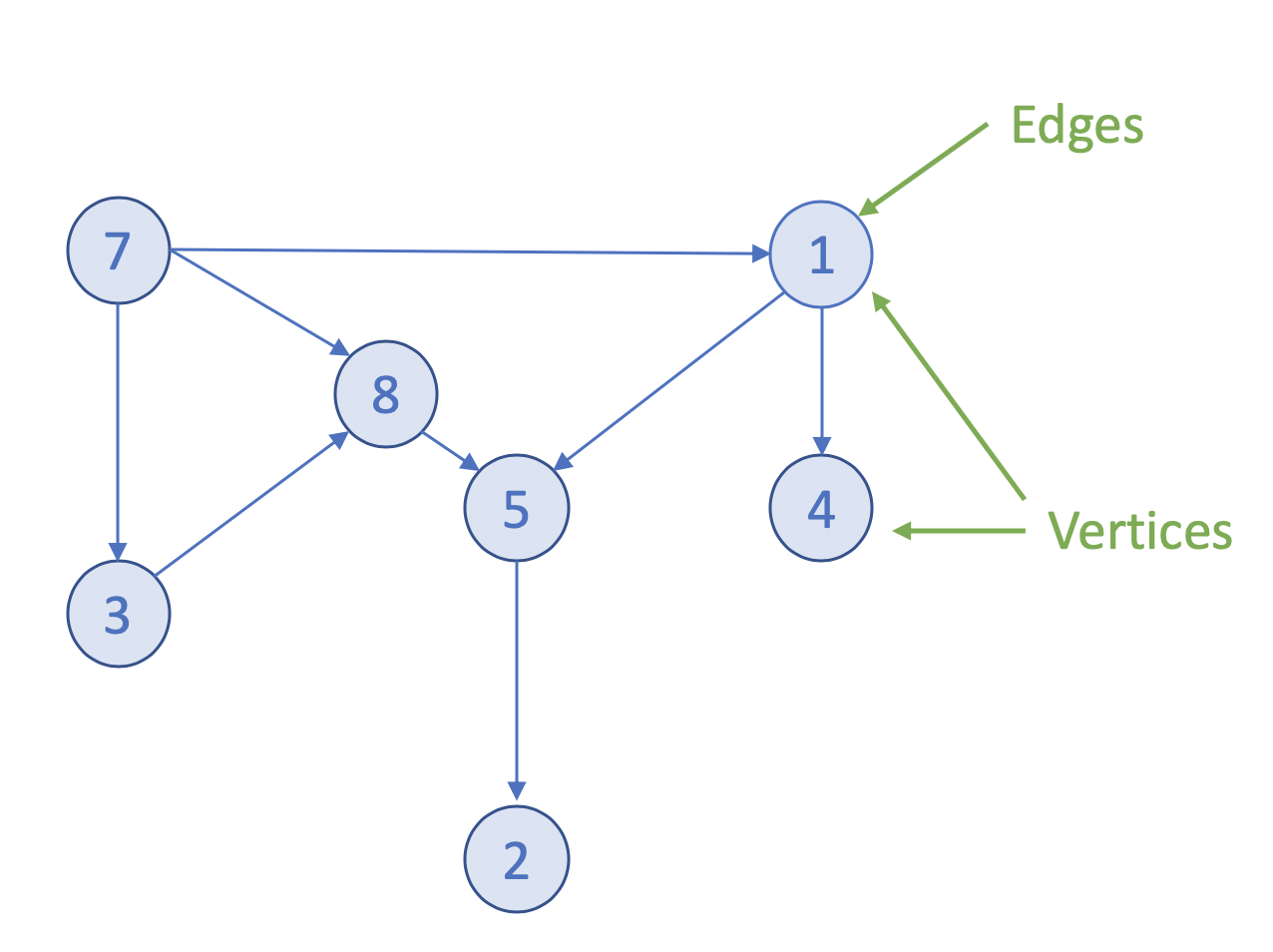
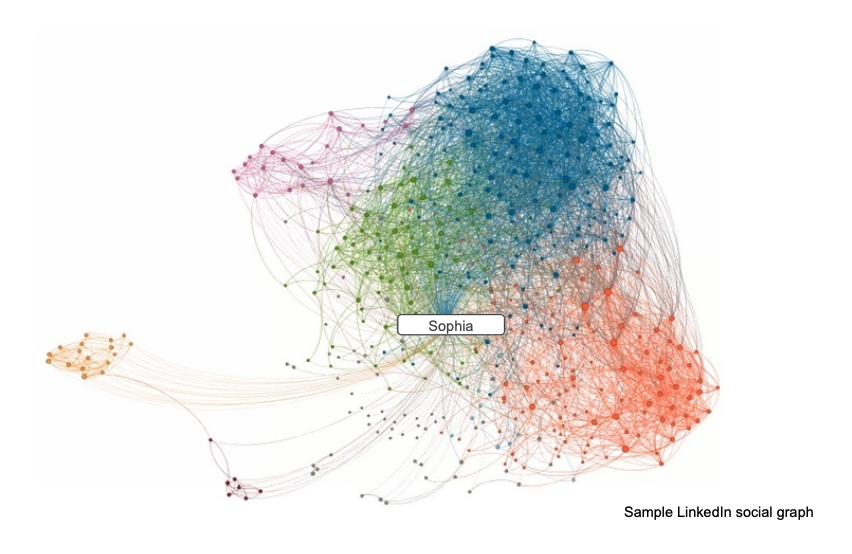
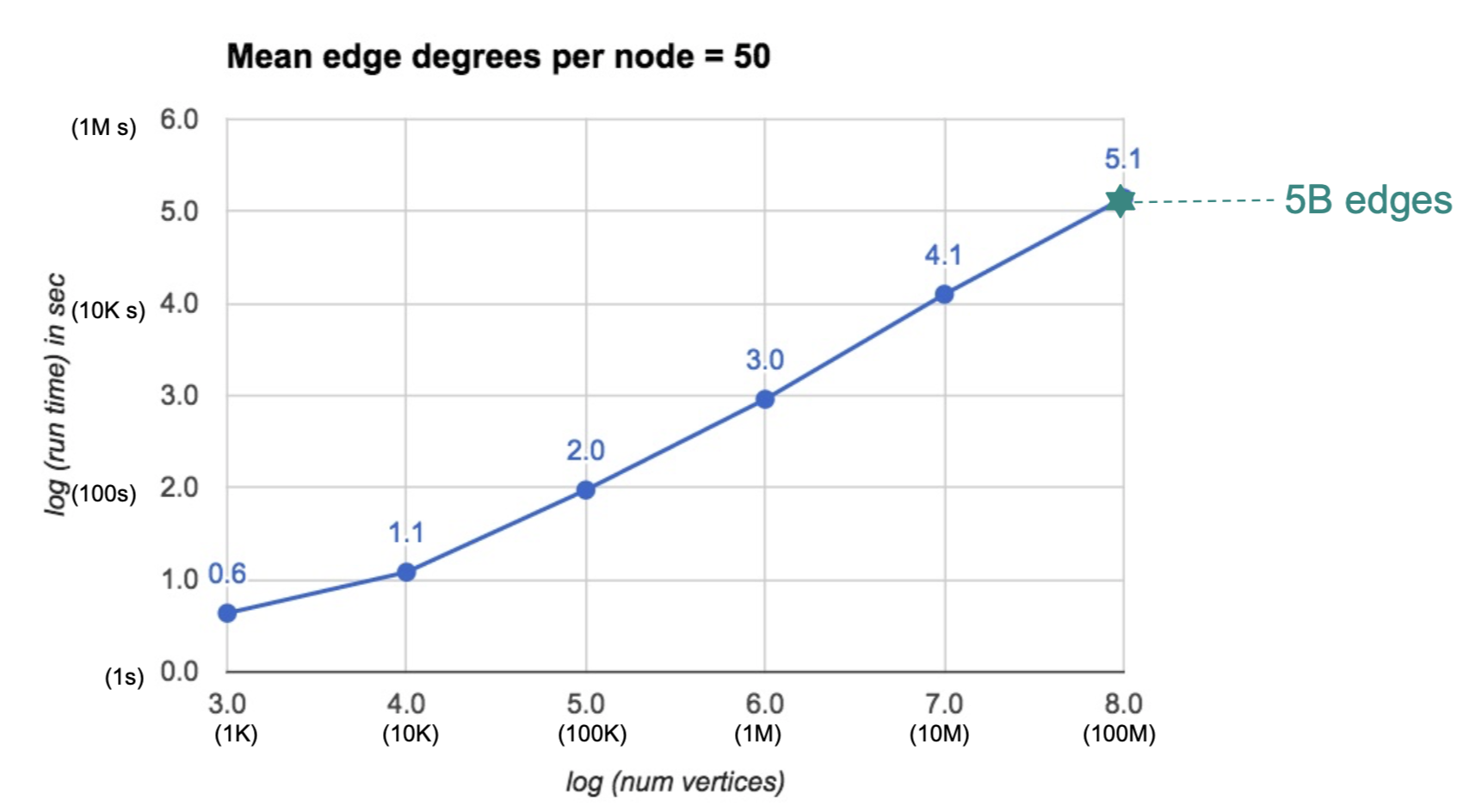
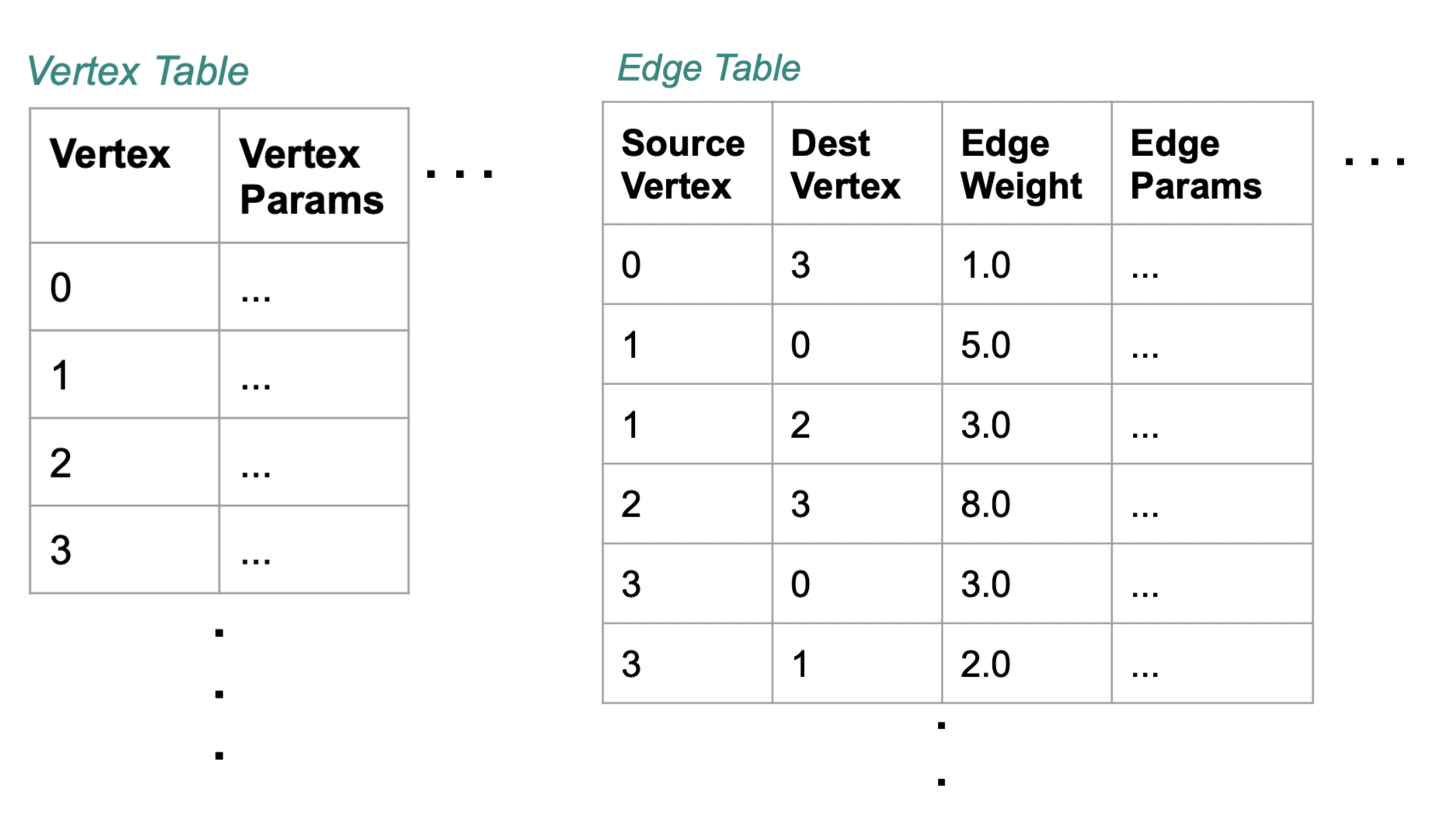
docs - graph analytics new page (#10138)
* clarifying pg_upgrade note * graph edits * graph analytics updates * menu edits and code spacing * graph further edits * insert links for modules
Showing
107.0 KB
166.0 KB
162.7 KB
473.0 KB
333.7 KB
381.7 KB
49.0 KB